Introduction
It has been previously reported
that the gluten metabolizing bacteria in the oral biofilm are involved in the
digestion and processing of gluten containing food products, such as, Rothia
aeria and R. mucilaginosa. While the human digestive enzyme
system lacks the capacity to cleave the immunogenic gluten, such activities are
naturally present in the oral microbial enzyme repertoire. Therefore, the human
microbiome contributes significantly to the digestion of food, especially the oral
microbiome [1,2]. The oral microbiome is very rich in microbial species, with
over 1000 oral taxa so far identified. The estimated numbers of bacteria in
dental plaque and saliva are 1011 per gram of dental plaque and 108
per ml of saliva, making the oral cavity the second most densely colonized part
of the human digestive tract after the colon. In addition, saliva contains a
wide variety of species that differ distinctly from the communities in the gut [3-8].
It has been published that the oral
microbiome is a novel and rich source of gluten degrading enzymes originating
from the oral microbiome. This is quite important because mammalian digestive
enzymes are reportedly only partly capable of cleaving gluten, and fragments
remaining induce toxic responses in celiac predisposed individuals. The effect
of the microbiome on individuals, with and without Celiac
Diseases (CD) was reported by Cameron demonstrating the role of gluten
metabolizing bacteria [9-11].
In that study the role of the genus Lactobacilli was strongly implicated. In another recently reported study, four strains from the species Lactobacillus ruminis, Lactobacillus johnsonii, Lactobacillus amylovorus and Lactobacillus salivarius, isolated from the proximal gastro-intestinal tract showed the highest peptide-degrading activities.
These strains displayed different
degradation rates and cleavage patterns that resulted in the reduction but not
the complete removal of immunogenic epitopes. This underscores the importance
of the human microbiome in digestion of food [12].
In addition, it is estimated that
over one liter of saliva is swallowed every day, taking the oral microbiome
into the proximal gastro-intestinal tract, affecting digestion and the
gastro-intestinal microbial constituency. A new area of study, the effects of
bacteria on the metabolic end-products, and the effects of the metabolome on
genetic expression, referred to as epigenetics, further emphasizes the
importance of studying the oral microbiome. Probiotic bacteria to remedy gluten
sensitivity have been recommended and clinical trials are progressing. What is
interesting is that many of these suggested probiotics for CD are already
probiotics commercially available [13-15].
Previous studies implied the
importance of these bacteria for societies consuming the modern “western” diet.
Also present in the modern “western” society is a reported increase in Irritable
Bowel Disease due to an alteration in the gut microbiome. In addition,
western culture also emphasizes the use of oral medicaments, ostensibly to
promote oral health. Over The Counter products may alter the oral microbiome
creating a situation less conducive for the survival of essential beneficial bacteria
[16-19].
Indeed, the uses of OTC oral
mouth rinses have been linked to high blood pressure, erectile dysfunction, low
capillary re-perfusion, diabetes, and obesity. It is postulated that the use of
OTC products may decrease the enzymatic
degradation of gluten containing foods by Rothia bacteria resulting in gluten sensitivity, Irritable
Bowel Syndrome (by the resultant shift in the gut microbiome), and
exacerbating ulcerative colitis increasing Celiac disease clinical prevalence.
In a previous research study, some of these oral medicaments were determined to
greatly inhibit the gluten metabolizers in
vitro. Therefore, the importance of the gluten metabolizing bacteria should
not be minimized and deserves further investigation into why some people have a
decreased level of these essential probiotics. The literature also does not
report how commonly the gluten metabolizing bacteria are present in our
environment and in the oral cavity of other mammals [20-23].
Objective
To isolate previously
undiscovered gluten metabolizing bacterial species from environmental sources
and to determine the factors, such as Over the Counter mouth rinses and
antagonistic bacteria responsible for their inhibition.
Materials and Methods
Previously non-investigated
sources of bacteria capable of digesting gluten were determined and the sites
cultured with swabbing using the Amies collection media. The sites were
commonly found areas where grain was reduced to flour, such as, grain mills,
and bakeries using non-bleached flour. The animal sources included common
household pets. The collected samples were incubated on gluten agar and the
colonies isolated. Colonies of bacteria growing on gluten agar were
sub-cultured to fresh gluten agar to confirm gluten utilization. Putative
gluten utilizing bacterial were then identified to genus and species level by
standard laboratory methods [24].
Susceptibility
Experiment
The inhibitory effects of various
oral mouth wash and other oral preparations were tested using a Kirby-Bauer
type assay. Oral bacteria of interest were grown in Mueller-Hinton media to a
McFarland Standard of 0.5. Trypticase Soy agar plates with 5% sheep blood were wholly
spread with one cotton swab inoculation of chosen bacteria to create a
bacterial lawn. Blank cotton discs were evenly distributed on the plate and 10
or 20 microliters of full strength test substrate was pipetted directly onto
each corresponding disc. Gluten metabolizing bacteria were challenged with
previously investigated known oral medicaments that inhibit the Rothia genus. The plates were evaluated
after 30 hours of growth at 36oC.
Calipers were used to measure zones of inhibition in millimeters.
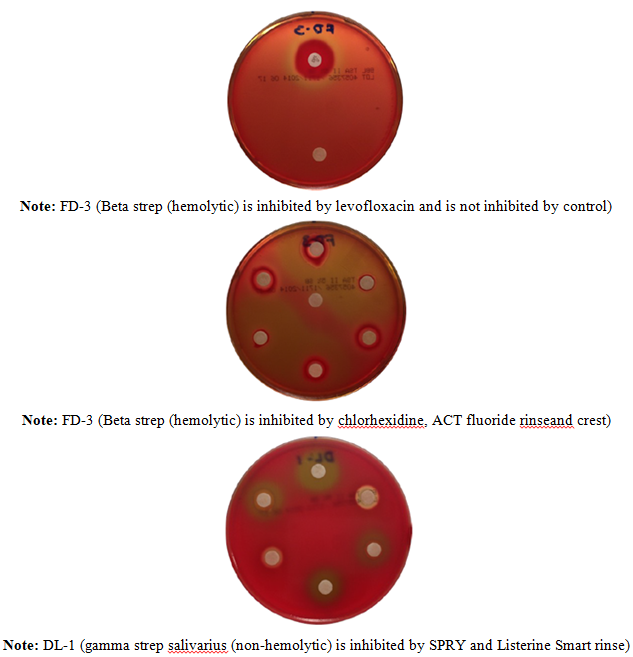
Bacteriocin
Detection Studies
Trypticase Soy Agar (TSA) was
autoclaved and cooled to 56 degrees and aliquots of 25mL were cooled and
inoculated with 2mL of 0.5 McFarland Standard suspensions of the experimental
bacteria prior to pouring agar plates. Impregnated plates were then inoculated
in punched zones using a disposable 10 microliter pipet with 0.5 McFarland
Standards of bacteria species: Streptococcus
salivarius, Staphylococcus aureus,
Vancomycin-resistant Enterococcus, Pseudomonas
aeruginosa, Escherichia coli, and
R. dentocariosa or R. mucilaginosa. The plates were
evaluated after 24 hours of growth at 36oC. Calipers were used to
measure zones of inhibition. Bacteriocin assays were also performed on the Rothia species, and the newly isolated
gluten metabolizers, MLC 124, LJ 514, AM 419 and BK as target strains.
Results
Oral medicaments such as CrestTM,
ListerineTM, ActTM Fluoride rinse, Chlorhexidine, and
Smart RinseTM inhibited all 16 of the new
gluten metabolizing bacterial strains (average 10 mms.). One strain MLC 124
was more resistant to oral medicaments. Xylitol products only inhibited 9
strains, but not MLC 124. Forty isolates were screened for bacteriocin activity
with Rothia species and the newly
isolated bacteria as targets. No zones of inhibition were detected with strain
identified as MLC 124. The 15 Factor a Groups demonstrated significant
differences as to Sensitivity to Oral Medicaments (DF 14, P=0.0005). The
following groups presented with significant differences (Bonferroni pair
testing): A1 vs B2, B1 vs B2, A1 vs B3, B1 vs B3, B3 vs B5, B3 vs B6, B2 vs B5,
and B2 vs B6.
Discussion
Nonpathogenic environmental and
non-human gluten metabolizing bacteria may prove beneficial as a source of
probiotic strains. Further identification of potentially beneficial bacteria is
recommended and may be of great importance. This study examined additional microbial
strains that were determined to already be present in the human oral
microflora, but were never previously considered to have any identifiable
purpose. Additionally, use of anti-microbial products appears to have a more
global influence than may have been believed. Although great caution should be
used when interpreting in vitro laboratory data into clinically relevant
results, pilot studies into the effects of oral medicaments should increase
further research efforts investigating these potential issues
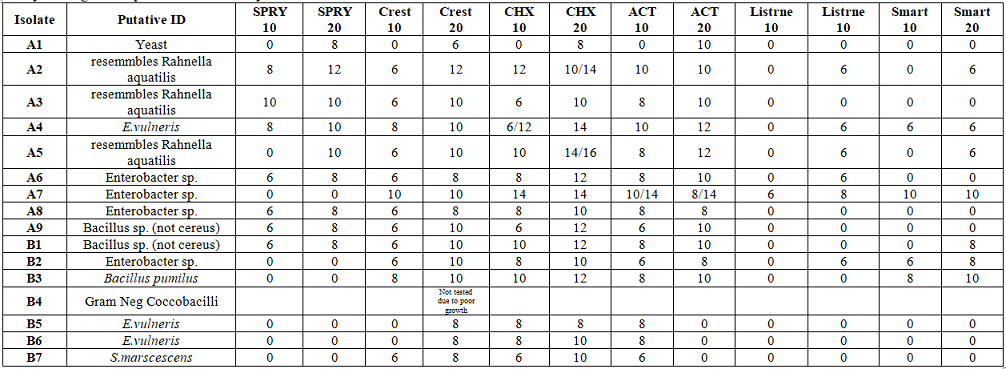
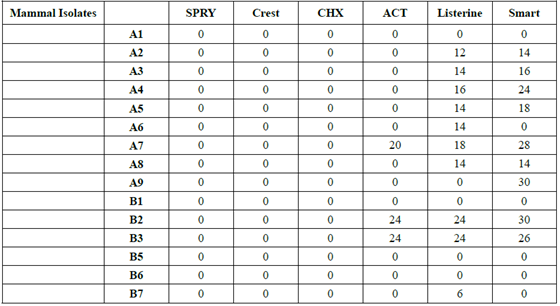
Table 2:Isolates from non-human sources inhibition by oral medicaments.
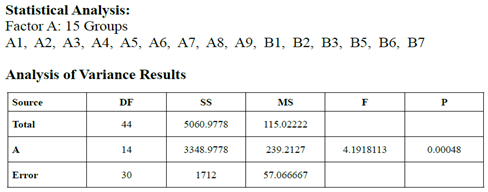
A statistically significant
result was demonstrated within and between the 15 bacterial strains as to
sensitivity to oral medicaments. This should indicate that certain bacterial
strains are more resistant to oral medicaments and would possibly be beneficial
as gluten metabolizing probiotic strains without concern as to oral hygiene
products being used by our patients.
Further research into the role of
gluten metabolizing oral strains is warranted, as is the development of gluten
metabolizing probiotics. Oral health professionals should be concerned over the
overuse of antimicrobials
in hygiene products, not just because of the effects on the many beneficial
oral bacteria but also because of possible gastro-intestinal strain shifting. This
may, possibly even create epigenetic events in the host. As a result,
antimicrobial oral hygiene products should only be utilized under the direct
supervision of a dental specialist. In theory, all medications should be
screened on an individual basis not only for appropriateness and efficacy, but
possible untoward sequelae.
Discovery of additional gluten
metabolizing bacterial species should be continued with emphasis on finding
strains that are resistant to antibiotics, microbial antagonists and over the
counter products. Ideally, these gluten metabolizers would also be beneficial
probiotics, inhibiting pathogens and positively modulating the host immune
response. Specifically because humans have not evolved to properly manage the
significant changes to our diet and environment, especially since the start of
the Neolithic agricultural revolution. But apparently, our microbiome has
evolved to help accommodate our dietary “adventures”. Unfortunately, the more
recent “fast food” revolution, along with the great expansion of preserved
convenience food, has further challenged the human oral and gut microbiome by
reducing in quantity, many commensal and probiotic bacterial strains previously
found in the diet. An additional complication is the hygienic conditions now
used to prepare food. Grain that was ground into flour by an exposed stone
wheel had a rich abundance of naturally present gluten metabolizers. Not so in
present times as the flour facilities are kept exceedingly clean, and the flour
is most often bleached, killing off an essential source of the gluten
probiotics. Possibly all commercial flour should be fortified with gluten
metabolizers that are potent probiotics. Further research should be performed
to test the limits of this proposed solution.
Household pets were not a
significant source of gluten metabolizers, and some strains were inhibited by
OTC oral products. Household pets are often referred to as “facultative” or
“obligatory” carnivores, and as such, should not have gluten metabolizers as
significant contributors to their oral microbiome. Obviously, they should not
be fed high gluten pet food products.
Conclusion
Newly discovered bacterial strains capable of digesting gluten that are resistant to oral antimicrobial agents and antagonistic (bacteriocin producing) bacteria were isolated from flour “environments”.
References
- Wei G, Zamakchari M, Dewhirst F, Schuppan D, Oppenheim F, et al. Rothia Bacteria as Gluten-Degrading Natural Colonizers of the Oral Cavity (2012) Presentation at the American Association of Dental Research Meeting, United States.
- Zamakhchari M, Wei G, Dewhirst F, Lee J, Schuppan D, et al. Identification of Rothia bacteria as gluten-degrading natural colonizers of the upper gastro-intestinal tract (2011) PLOS ONE 6: e24455. https://doi.org/10.1371/journal.pone.0024455
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, et al. The human oral microbiome (2010) J Bacteriol 192: 5002-5017. https://doi.org/10.1128/jb.00542-10
- Maukonen J, Motto J, Suihko ML and Saarela M. Intra-individual diversity and similarity of salivary and faecal microbiota (2008) J Med Microbiol 57: 1560-1568. https://doi.org/10.1099/jmm.0.47352-0
- Li Y, Ku CYS, Xu J, Saxena D and Caufield PW. Survey of oral microbial diversity using PCR-based denaturing gradient gel electrophoresis (2005) J Dent Res 84:559 -564. https://doi.org/10.1177/154405910508400614
- Nisengard RJ, Newman MG. 1994. Oral microbiology and immunology, 2nd ed. Saunders, Philadelphia, PA.
- Ritari J, Salojarvi J, Lahti L and de Vos WM. Improved taxonomic assignment of human intestinal 16S rRNA sequences by a dedicated reference database (2015) BMC Genomics 16:1056. https://doi.org/10.1186/s12864-015-2265-y
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, et al. Bacterial community variation in human body habitats across space and time (2009) Science 326: 1694-1697. https://doi.org/10.1126/science.1177486
- Fernandez-Feo M, Wei G, Blumenkranz G, Dewhirst FE, Schuppan D, et al. The cultivable human oral gluten-degrading microbiome and its potential implications in coeliac disease and gluten sensitivity (2013) Clinical microbiol infec 19: E386–E394. https://doi.org/10.1111/1469-0691.12249
- Helmerhorst EJ, Zamakhchari M, Schuppan D and Oppenheim FG. Discovery of a novel and rich source of gluten-degrading microbial enzymes in the oral cavity (2010) PLoS One 5: e13264. https://doi.org/10.1371/journal.pone.0013264
- Caminero A, Nistal E, Herran AR, Perez-Andres J, Ferrero MA, et al. Differences in gluten metabolism among healthy volunteers, coeliac disease patients and first-degree relatives (2015) Br J Nutr 114: 1157-1167. https://doi.org/10.1017/s0007114515002767
- Francavilla R, De Angelis M, Rizzello CG, Cavallo N, Dal Bello F, et al. Selected probiotic lactobacilli have the capacity to hydrolyze gluten peptides during simulated gastrointestinal digestion (2017) Appl environ microbiol 83: e00376-17. https://doi.org/10.1128/aem.00376-17
- Tan S, Liang CR, Yeoh KG, So J, Hew CL, et al. Gastrointestinal fluids proteomics (2007) Proteomics Clin Appl 1: 820-0833. https://doi.org/10.1002/prca.200700169
- Aleksandrova K, Romero-Mosquera B and Hernandez V. Diet, gut microbiome and epigenetics: emerging links with inflammatory bowel diseases and prospects for management and prevention (2017) Nutrients 9: 962. https://doi.org/10.3390/nu9090962
- Chander AM, Yadav H, Jain S, Bhadada SK and Dhawan DK. Cross-talk between gluten, intestinal microbiota and intestinal mucosa in celiac disease: recent advances and basis of autoimmunity (2018) Frontiers microbial 9: 2597. https://doi.org/10.3389/fmicb.2018.02597
- Amato KR, Yeoman CJ, Cerda G, Schmitt CA, Cramer JD, et al. Variable responses of human and non-human primate gut microbiomes to a Western diet (2015) Microbiome 3: 53. https://doi.org/10.1186/s40168-015-0120-7
- Gomez A, Sharma AK, Mallott EK, Petrzelkova KJ, Jost Robinson CA, et al. Plasticity in the human gut microbiome defies evolutionary constraints (2019) mSphere 4: e00271-19. https://doi.org/10.1128/msphere.00271-19
- Distrutti E, Monaldi L, Ricci P and Fiorucci S. Gut microbiota role in irritable bowel syndrome: New therapeutic strategies (2016) World J gastroenterol 22: 2219-2241. https://doi.org/10.3748/wjg.v22.i7.2219
- Joshipura KJ, Muñoz-Torres FJ, Morou-Bermudez E and Patel RP. Over-the-counter mouthwash use and risk of pre-diabetes/diabetes (2017) Nitric oxide: biol chem 71: 14-20. https://doi.org/10.1016/j.niox.2017.09.004
- Hyde ER, Andrade F, Vaksman Z, Parthasarathy K, Jiang H, et al. Metagenomic analysis of nitrate-reducing bacteria in the oral cavity: implications for nitric oxide homeostasis (2014) PloS one 9: e88645. https://doi.org/10.1371/journal.pone.0088645
- Koopman JE, Buijs MJ, Brandt BW, Keijser BJ, Crielaard W, et al. Nitrate and the origin of saliva influence composition and short chain fatty acid production of oral microcosms (2016) Micro ecol 72: 479-492. https://doi.org/10.1007/s00248-016-0775-z
- Kapil V, Haydar SM, Pearl V. Lundberg JO, Weitzberg E, et al. Physiological role for nitrate-reducing oral bacteria in blood pressure control (2013) Free radical boil med 55: 93-100. https://doi.org/10.1016/j.freeradbiomed.2012.11.013
- Cannon M, Muhammad A, Jantra L, Kabat W and Yogev. In vitro investigation of gluten metabolizing bacteria and their inhibition (2015) American Academy of Pediatrics, United States.
- Versalovic J, Carroll KC, Jorgensen JH, Funke G, Landry ML, et al. Manual of Clinical Microbiology (2011) ASM Press, United States.
*Corresponding author
Mark L Cannon, Professor, Division of Dentistry,
Department of Otolaryngology, Feinberg School of Medicine, Northwestern
University, Chicago, Illinois, USA, Tel: +847-899-6720, E-mail: drmarkcannon@outlook.com
Citation
Cannon M, Kabat B, Yogev R, Awan A, Jantra L, et al. Investigation into gluten metabolizing bacterial species and their inhibition (2020) Edel J Biomed Res Rev 2: 1-4.
Keywords
Gluten, Microbiome, Oral anti-microbial products.


 PDF
PDF
