Research Article :
Zong-Sian Chen,
Jean Trejaut, Jun-Hun Loo, Ying-Hui Lai,
Jin-Yuan Huang and Marie Lin This study investigates the genetic relationship
of the Mazu peoples on the east coast of China in the Taiwan Strait. Using
partial and complete mitochondrial DNA (mtDNA) sequences, we compare Mazu with
surrounding East Asia populations. Mazu shows no exclusive affinities with
either Southeast or Northeast Asia. High genetic diversity and a very high
number of exclusive haplogroups of various Asian origins suggest that Mazu
resulted from a process of continuous resettlement that started when it first
became an archipelago at the end of the last glacial maximum and that continued
till the last century. As a result, genetic drift did not contribute to an
exclusive Mazu profile. The structure of haplogroups that show signatures of
the Neolithic era (N9a10a), or influx from Island Southeast Asia (F1a4a)
suggest recent gene flows and Mazu relationship with it's pre-Neolithic era
(presence of pre-E1a or R9/pre-F from Liangdao man) was not seen. NanGan island, in the Taiwan
Strait of the China Sea, is part of a 36 island and islets archipelago. The
main islands are not all inhabited and comprise Hsijiu, Tungjiu, Nangan, Beigan,
Gaudeng, Dachiu, Hsiaochiu, Liangdau, Shiyin, and Tungyin. Along with Penghu
and Kinmen islands further south in the Taiwan
Strait, the Mazu archipelago is a separate customs territory of Taiwan and is
administrated by the Lienchiang County of Taiwan, (also termed Lianjiang, and
often referred to Mazu County) (https://www.cia.gov/library/publications/the-world-factbook).
Following the establishment of
garrisons on some islands, the population
rapidly numbered to 17000 [1]. Like urban people in Taiwan, most natives of
Mazu believe they share their origin with Northern Fujian and Fuzhou peoples
who migrated to Taiwan in the last 400 years. The finding of large shell mounds
with radio-carbon
dating ranging from 8000 to 4000 years BP showed that a hunter-gatherer
culture, with coastal foraging lifestyle, inhabited Mazu Islands [2]. More
recently, a study combining ancient DNA (aDNA) from two 8000 years BP human
skeletons in Liang island (nearby island to Nangan; (Figure 1) and complete mtDNA genome sequences representing modern
Taiwan indigenous and non-indigenous peoples showed these skeletons belonged to
mitochondrial
haplogroups E1 and R9/F and proposed they were ancestors of the Formosan
indigenous peoples, predating Taiwan Neolithic era of demographic expansion
6,000 BP, and likely had been speakers of proto-Austronesian [3]. During the Yuan Dynasty at the
beginning of the XIIIe century, fishermen from nearby Fujian and Zhejiang on
the east coast of China
used the islands for shelter while few eventually settled there permanently.
But starting In the XVII century and during the entire length of the Qing
Dynasty, the east coast of China became a very frequented maritime highway.
Because of these successive events, fishermen, traders, sea nomads, and pirates
may have successively contributed to displacing the first or former residents
[2]. Present-day Mazu peoples use a
Sinitic sub dialect spoken in the nearby Chinese city of Fuzhou. A minority of
individuals with a sea-dwelling mode of living use different dialects thought
to be related to Mainland Southeast Asia (MSEA) populations [4]. However,
Mandarin Chinese has now become the official language. Because of its
administrative relationship with Taiwan, and its pre-Neolithic heritage,
the genetic diversity of the Mazu people is important to an understanding its
genetic relationship with Taiwan Han (TwH), Taiwan Indigenous People (TWIP),
Northeast Asia (NEA), East and Southeast Asia (EA and SEA), Mainland Southeast
Asia (MSEA) and Island Southeast Asia (ISEA) [5-7]. In this study,
Mitochondrial DNA (mtDNA) is used to characterize the genetic diversity and
characterize the relationship of the Mazu peoples with Asian populations of
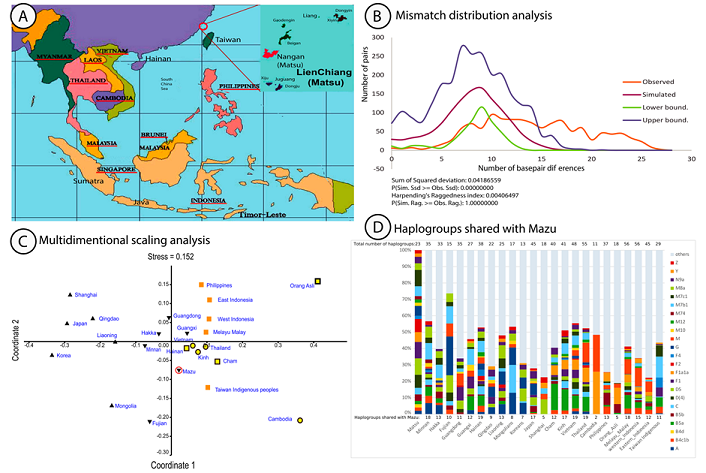
distinct languages and cultures. Figure1: A) Map of Southeast Asia and the Mazu archipelago. B) Pairwise Mismatch Distribution A subset of 50 mouth swabs of unrelated
Mazu individuals from Nangan island (Mazu) was analyzed by sequencing for
mitochondrial DNA diversity using coding region (Nucleotide Positions (nps)
8000 to 9000 and nps 9800 to 10873), and the control region of the
Hyper-Variable Segment II (HVSII) nps 16030 to 16410. This totaled to 2453 loci
of which 116 were polymorphic. Ambiguous haplogroups assignments using the
HaploGrep version 2 software were confirmed using further determination of
pertinent SNPs of the coding region [7,8]. For the distribution analyses, the
genetic data for current populations from North and South Asia, Southeast Asia
and Mainland Southeast Asia were retrieved from published reports. Statistical
analyses and pairwise genetic distances of Rst between different
populations were carried out using the Arlequin software 3.1 and visualized
using the MDS tool available in the PAST software [9,10]. the Network software
[11]. Out of 38 exclusive mtDNA haplotypes
observed among the 50 Mazu individuals, 29 haplogroups
were seen only once. At a lower assignment level eight basal phylogenetic
haplogroups lineages (D4, D5, F1, F2, G and M7) had a frequency greater than 5%
and represented 64% of the total diversity. Among them, sub-types of D4, D5, F2
and G prevail in modern populations of NEA, while Sub-types of F1 (F1a1’,
F1a1a, F1a1c, F1a4a and F1c1a1) and M7 (M7c1’, M7c1b2b and M7c1c2) are rarely
seen in Northeast Asia and prevailed in SEA and MSEA where they likely
originated [12,13]. M8a2'3, M10a1+16129, M12a1a1, M74. Other haplogroup that
are common in MSEA were M8a2'3, M10a1+16129, M12a1a1, M74. B4d1 seen only once
was unique in Mazu. Although Northeast Asian (Japan 77.8% and Liaoning 51.9%)
and Taiwan Han (Minnan 55.6% and Hakka (51.9%) are the highest contributor of
haplogroups to Mazu. The specific relationship of Mazu with
Taiwan was minimal. Using our raw data level of haplogroups assignment only
haplogroups, B4c1b2a and F1a4a’, showed similar patterns of distribution in
insular East Asia, with low prevalence among Taiwan Han but common among
Formosan indigenous people and the Philippines.
Except for the low occurrence of F1c1a1 and N9a1'3 among Taiwan Han and Taiwan
Pingpu none of the other Mazu haplogroups showed prevalence in Taiwan. The heterogeneous profile described
above was supported by a high haplogroups diversity (h) (h=0.988; SE 0.003) and
a nucleotide diversity (average over loci) of 0.005763 +/- 0.002916.
Interestingly the tests of neutrality of Tajima’s D and Fu’s Fs (Schneider et
al., 2000) [9] showed negative values (D=-1.700; p=0.019 and Fs=-24.32
p<0.0001 respectively), suggesting population expansion. However, the high
haplogroup diversity, the high number of single haplogroups presume a
relatively high number of unique
nucleotide variations, are indicative of an ethnically heterogeneous
population, and are the result of numerous and separate gene flow from various
regions of East Asia that likely started in the Neolithic era until the present
days [14,15]. In support to this observation, the
mismatch distribution analysis (Figure 1B) showed a flat and wide uni-modal
observed curve (in red in Figure 1B) between 5 and 25 base-pair differences
(bp) and a bp mean of 14.5. In contrast with Tajima’s D and Fu’s Fs tests, we
here reject the hypothesis of sudden expansion, this is shown by a significant
Sum of Squared Deviation (SSD) test (SSD=0.042. p<0.0001), and a low value
of Harpending’s raggedness (r=0.0041, p=0.95) (Figure 1 B) indicating a robust
analysis and are the result of mixtures of groups of many Asian origins rather
than sudden population
expansion [16]. Multidimensional Scaling analysis using
FST statistic, a measure of population substructure comparisons (Figure 1C),
was used to test the matrilineal spatial genetic distribution between Mazu and
other Asian populations (Figure 1C). The distribution plot (stress: 0.152)
differentiated clearly NEA and SEA Chinese groups (Black) from ISEA groups
(Orange) and MSEA (Yellow). These ethnic clusters also associated well with the
distribution of Sinitic, Austronesian, and Austroasiatic languages (triangles,
squares, and circles respectively). Low FST values were seen between Mazu and
Hainan, or Mazu and Vietnam, however, both FST values were significant (FST 0.02,
p< 0.0001, and FST 0.007, p=0.72 respectively) suggesting distant
relationships. Interestingly, all FST values between Mazu and other populations
of NEA, SEA, MSEA, ISEA, Taiwan Han and Taiwan Indigenous people showed
significant FST values (p< 0.001) (data not shown) or relationship of the
past. These observations
support the very high level of genetic differentiation described above, and the
likely probability that Mazu was never settled for a long time by the same
group. To this date, no archaeological traces
indicating the passage of anatomically modern humans in Mazu during the
Pleistocene (40,000 to pre-Holocene) [3]. The first incursion modern human in
the Mazu archipelago is dated in early Holocene on Liang island (herein
referred to as Mazu) most likely already an Island by 8,000 YBP. Marking this
period, ancient human mtDNA remains have revealed haplogroups R9/pre-F and a
precursor of E1a, both of southeast China origin [3]. These haplogroups have no descendant
ramifications, but sister descendants seen among Austronesian speakers
of Taiwan and the ISEA. To clarify the genetic relationships of modern Mazu
with a pre-Neolithic past, we collected mtDNA genome data from 50 unrelated
Mazu individuals. When compared with other populations of East Asia, the
haplogroups profile showed population differentiation to an extent not expected
in present-day East Asians Islanders, and revealed a complex population
history. The mid-Neolithic period to the start of
the first millennium of our era went through the start of food globalization
and the spread of northern East Asians toward southern East Asia which affected
the genetic ancestry of southern China [4]. Similarly, movements of coastal groups
such as migrants, traders, Pirates, and sea nomads from MSEA to North Asia and
back suggests that gene flow also played an important role in the
prehistorically genetic makeup of coastal East Asia and Mazu. These population
movements continue throughout this era, up to the last century, with Mazu
archipelago becoming a temporary refuge to local fishermen. The introduction of
more recent settlers and the establishment of garrison from Taiwan in the last
century after Mazu became administrated by Taiwan resulted in a major increase
of the Mazu population. The present-day genetic variation is
characterized by a very high polymorphism, with numerous mtDNA lineages (from
MSEA, NEA, SEA, and Taiwan Han). None of these phylogenetic lineages are deeply
related by descent, indicating a shallow to no evolutionary time and recent
series of temporary settlements of Mazu. In general, all these people signed
their passage into the genetic structure of Mazu with a slightly more marked
Northeast Asian than Southeast Asian/MSEA influence, with little gene
flow from the Taiwan Indigenous people. Finally, No traces of descent of
R9/pre-F and pre-E1a from the original pre-Neolithic settlers were found in the
extant population. In summary, the maternal
inter-population comparisons reveal that the Mazu people have a similar affinity
with northern Asian populations (Chinese, Japan, Korea) and the Southern Asian
Population (SEA and MSEA; 48%). Affinities with Austronesian speaking groups
were rare. Bar-chart of haplogroups sharing suggested important
multi-regional gene flows and along with the MDS, analysis supported a pattern
of admixture reflecting the complex settlement process of Coastal East China.
All traces of descent of
R9/pre-F and pre-E1a from the original pre-Neolithic settlers were diluted in
the extant population. We thank the Ministry of Science and
Technology of Taiwan for making this project possible. We are also grateful to
the people of Taiwan for donating their blood. No benefits in any form have been received
or will be received from a commercial party related directly or indirectly to
the subject of this article. The authors have declared that no competing
interests exist. This work was supported by grant
No.106-2320-B-195-001-MY2 from Ministry of Science and Technology of Taiwan. Marie
Lin, Molecular Anthropology and Transfusion Medicine
Research Laboratory, Mackay Memorial Hospital, Taipei, Taiwan, Tel:
+886-2-2809-4661#2380, FAX: +886-2-2809-8746, E-mail: marielin0530@gmail.com
Chen ZS, Trejaut JA, Loo JH, Lai YH, Huang JY, et al. mitochondrial DNA diversity of the nangan
islanders living in the mazu archipelago of the Taiwan strait (2021) Edel J Biomed Res Rev 3: 25-27 Mazu Population, Mitochondrial DNA Inheritance,
Haplotype sharing with Taiwan and surrounding East Asia populations.Mitochondrial DNA Diversity of the Nangan Islanders Living in the Mazu Archipelago of the Taiwan Strait
Abstract
Full-Text
Introduction
Material
and Method
Results
and Discussion
Summary
Acknowledgements
Competing Interests
Funding
References
Corresponding
author
Citation
Keywords