Research Article :
Ghazay F Alotaibi and Mamdouh A Bukhari Microbes generally attach to surfaces and produce an Extracellular Polymeric Substance (EPS) matrix. The exopolysaccharide production plays a role in biofilm protection against environmental stress factors. Biofilm-forming bacteria exhibit different physiological properties in their response to environmental influences compared with their planktonic counterparts. This study aimed to investigate the impact of changing the concentrations of glucose, peptone, and yeast extract and environmental parameters, such as temperature, pH, anaerobic conditions, osmotic stresses, and growth media on biofilm formation by K. pneumonia MBB9 recovered from river-stones collected from the Porter Brook, Sheffield using crystal violet and resazurin assays in microtiter plates. The different concentrations of glucose (0.25, 0.5 and 1 g L-1), peptone (0.25, 0.5 and 1 g L-1) and yeast extract (0.25, 0.5 and 1 g L-1) as carbon and nitrogen sources found to have an impact on biofilm formation by K. pneumonia MBB9. The greatest biomass level being at 0.25 g L-1 for glucose whereas the density of biofilm increased significantly with increasing the concentration of peptone and yeast extract until 1 g L-1 of peptone and yeast extract, suggesting that higher levels of peptone and yeast extract can be beneficial for biofilm formation by K. pneumonia MBB9 in microtiter plates. The amount of biofilm was high at pH 4.5 and 0.6% NaCl; however, the significant reduction at pH 10.5 and 10.6% NaCl could be as a result of the slow growth under higher NaCl concentrations and highly alkaline condition. High-density biofilm produced at 40 °C; however, a temperature of 50 °C reduced the amount of biofilm by K. pneumonia MBB9, suggesting that more extreme temperatures might affect the formation of biofilm by inhibiting growth. Besides, biofilm production under anaerobic conditions was significantly lower (83% less) than under aerobic environments. Klebsiella pneumonia MBB9 possessed a high capacity to form biofilms on the surface of glass slide coupons. Bacterial
biofilms can be identified as microbial communities adhering to abiotic and
biotic surfaces and forming a secure mode of growth of the extracellular matrix
[1-3]. The matrix of biofilm typically guards microbes against external
conditions, such as pH variations, high salinity, pressure, depletion of
nutrients, oxygen radicals, antibiotics and disinfectants [4,5]. Biofilm
formation on different surfaces can be through different steps, including the
free floating planktonic
cells adhesion, maturation, and attached cell dispersion [6,7]. Establishing and
developing bacterial biofilm is considered a dynamic and complex process
controlled by inherent biological characteristics and also by several
environmental factors as variations in the environment usually trigger the biofilm
formation [8,9].The environmental factors controlling biofilm attachment
and formation can also affect bacteria's ability to grow and survive [10]. Many
factors, such as incubation time, nutrient concentrations, pH, temperature, and
ionic strength can influence the formation of bacterial biofilms; however,
bacterial cell surface appendages and surface properties are also required for
this process [11]. Surface attachment biofilm formation is governed primarily
through electrostatic, hydrophobic, van der Waals, and contact communications
[12,13]. Hydrophobic interactions play a role in bacterial adhesion to
different surfaces and promote biofilm formation. Surface hydrophobicity
can affect microbial material colonization since bacteria have developed
different ways to adhere to the substrate using the hydrophobic force. However,
the bacterial physiological state can change the hydrophobicity of the cell
surface [14-16]. Bacteria form
biofilms have many physiological properties compared to their counterparts in
suspensions in their response to environmental impacts; thus, change in
necessary nutrients availability can affect microbial physiology [17]. Temperature
plays an important role in regulating the bacterial activity rate and the
proliferation of biofilm and organism settlements in aquatic environments [18].
However, the biofilm accumulation rate can be increased by increasing carbon
levels [19]. An increase in water temperature, nutrients levels and flow
velocity may enhance the rate of bacterial attachment, provided that the
critical values of these factors are not exceeded [20]. The present study
aimed to examine the impact of changing the levels of glucose,
peptone and yeast extract and environmental parameters, such as pH,
temperature, osmotic stresses, temperature and growth media on K. pneumoniae
MBB9 biofilm formation using crystal violet and resazurin (also known as the
Alamar Blue) assays in microtiter plates.
Source and Sampling
Isolation of bacteria
from environmental biofilms Bacterial
morphological and biochemical characterization Molecular
identification of bacteria via 16S rRNA gene sequencing and phylogenetic
analysis SYBR Safe® (Invitrogen) was used to stain the gels. A UVI
tech photo documentation system was used to visualize DNA bands for viewing the
DNA fragments. Following the sequencing, each DNA sequence chromatogram
was examined using the bioinformatics tool Finch TV software to evaluate its
quality; however, sequences with low quality were trimmed from both ends and
moved the remaining good-quality sequences into a new file. Basic Local
Alignment Search Tool (BLAST) in National Center for Biotechnology Information
(NCBI) and Ribosomal Database Project (RDP) were used to produce taxonomic
information about the source species. The neighbor-joining method implemented
in the program MEGA software was used for construction of phylogenetic
tree. Assessment of biofilm
formation by the isolated bacteria using microtiter plate assay Planktonic cells in the fluid were then removed by inverting
the plate and decanting the contents, followed by thoroughly rinsing three
times with 200 μl of sterile deionised water (dH2O) to remove any remaining
unattached planktonic cells. The microtiter
plates were dried by air at 37 °C, and adherent bacteria were stained with 200
μl of 1% (w/v) crystal violet solution (crystal violet; Merck, Germany) for 25
min [25,26]. The supernatant was discarded after the staining step and the
wells were rinsed with repeated washing with sterile deionized water (dH2O) for
any excess stain removal. Any biofilm-integrated CV was solubilized by adding
250 μl of 30% glacial acetic acid. A multi-well plate reader (BioTek FLx800,
UK) at the absorbance of light at 595 nm was used to assess the CV liberated
from the attached material and control wells [27]. Determination of cell
activity of biofilm of Klebsiella pneumoniae MBB9 using resazurin Effect of glucose,
peptone and yeast extract on biofilm formation by Klebsiella pneumoniae MBB9 Effect of
environmental factors on biofilm formation by Klebsiella pneumoniae MBB9 Effect of temperature
on biofilm formation: The impact of temperature on biofilm formation was
assessed through incubating the inoculated 96-well plates under static
conditions at 25, 37, 40 and 50 °C for 24 h using crystal
violet and resazurin assays. Effect of anaerobic conditions
on biofilm formation: To evaluate the effect of anaerobic conditions on
biofilm formation by K. pneumoniae MBB9, microtiter plate cultures were
incubated anaerobically at 37 °C under anaerobic conditions for 24 h using
crystal violet and resazurin assays. Effect of osmotic
stress on biofilm formation: Nutrient broth medium adjusted to various NaCl
values was used to test the impact of pH on biofilm formation by K. pneumoniae
MBB9 in microplates using crystal violet and resazurin assays. Effect of different
growth media on biofilm formation: Nutrient
Broth (NB), Lysogeny Broth (LB) and Tryptic Soy Broth (TSB) were used to
assess the effect of growth media on biofilm formation by K. pneumoniae MBB9
using crystal violet and resazurin assays. Assessment of
biofilms formed by K. pneumoniae MBB9 on glass surfaces Quantification of
biofilms: At the end of each incubation period, a set of glass chips were
aseptically removed from the inoculated broth culture and rinsed three times with
sterile deionized water (dH2O) to remove unattached bacterial cells. Coupons
were then put into a testtube containing 40 ml of sterile deionized water
(dH2O) and sonicated at 20 Hz for 3 minutes. This mixture was homogenized by
vortexing in order to disperse the sessile cells from the chips' surfaces. Quantification
of biofilm cells was determined by direct count pour plate; suspensions
were serially diluted in sterile saline 0.85% w/v of normal saline, and the
plates of nutrient agar were inoculated with an aliquot (100 µl) of each
dilution and incubated at 37 °C for 24-48h. The number of cells was then
expressed as CFU per cm2 [30-32]. Fluorescent staining
and microscopical examination of live/dead cells: A Live/Dead BacLight
Bacterial Viability Kit (Eugene, Oregon, USA) was used according to the
manufacturer’s instructions to assess the biofilm cells on the glass slide coupons.
This set consists of a combination of SYTO® 9; the green fluorescent nucleic
acid stain and propidium iodide; the red-fluorescent nucleic acid stain
that can mark both live and dead bacteria [33,34]. In brief, 20 µl of
fluorescent stain was prepared as a working solution by incorporating 2 µl of
SYTO®9 stain with 498 µl of sterile deionized water (dH2O). The glass slide coupons
were flooded with 200 µl of the working solution, covered with aluminum foil
and incubated at room temperature for 15 minutes in the dark according to the
instructions of the manufacturer. Fluorescence microscopy was used to examine
the glass chips and to detect the fluorescence from SYTO®9 using a filter with
an excitation wavelength of 485 nm and an emission filter of 498 nm under 100X
oil immersion fluorescence objective. Fluorescence microscopy images were
analyzed using ImageJ software. Statistical analysis As described in my previous research, 22 different bacterial
strains were isolated and identified from biofilms formed on stones recovered
from the Porter Brook, Sheffield. Of the 22 isolates, ten gram-negative
potential pathogens were selected and screened for biofilm
production. The modified microtiter-plate test, as a quantitative assay,
showed that all tested strains produced biofilms as the mass of the retained
crystal violet stain on the test plate indicated the biofilms presence.
Klebsiella pneumoniae MBB9 were among these isolates that showed the highest
biofilm production in the CV microtiter plate assay [35]. Effect of glucose,
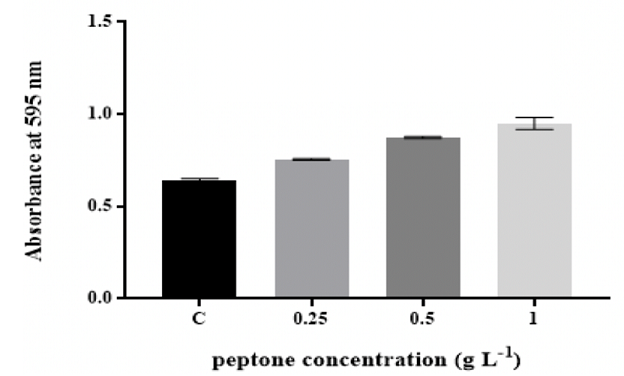
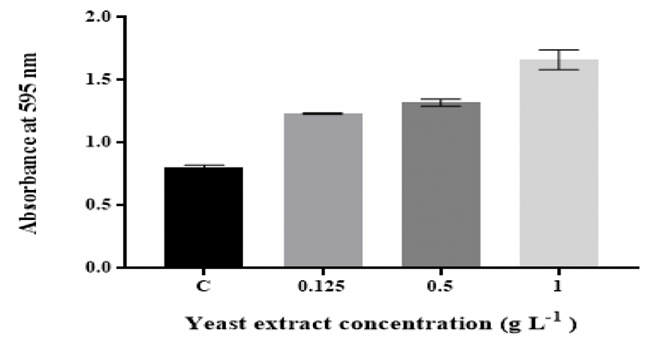
peptone and yeast extract Figure 1: Effect of D-glucose concentration on in vitro biofilm Figure 3: Effect of changing yeast extract concentration on in vitro
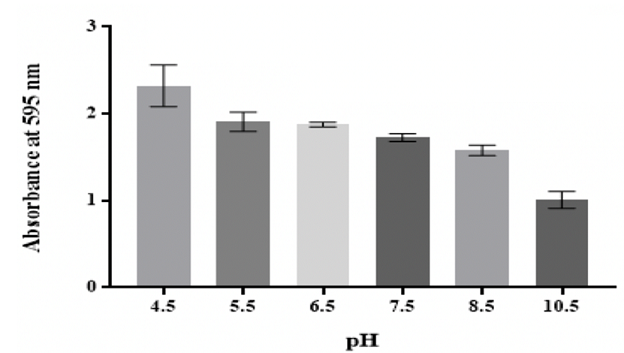
Effect of pH Figure 4: Effect of pH on in vitro biofilm formation by K. pneumoniae MBB9 Effect of temperature
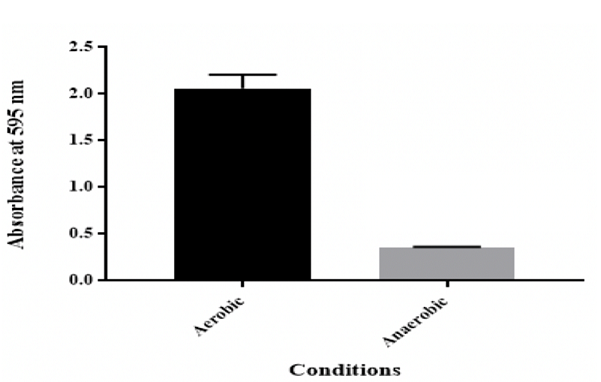
Figure 5: Effect of temperature on in vitro biofilm formation by K. Effect of anaerobic
conditions Figure 6: Effect of anaerobic conditions on in vitro biofilm formation by K. pneumoniae MBB9 Effect of osmotic
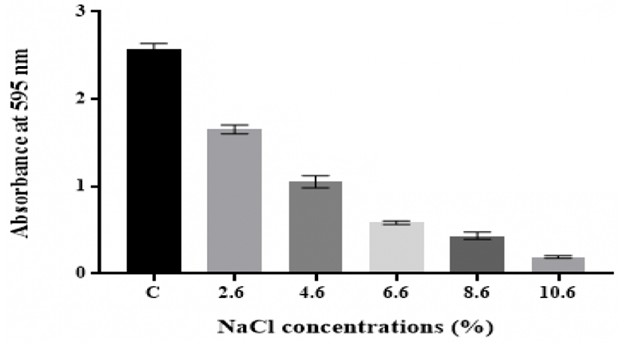
stress Figure 7: Effect of osmotic stress on in vitro biofilm formation by K. pneumoniae MBB9 Effect of different
growth media Figure 8: Effect of different growth media on in vitro biofilm formation by K. pneumoniae MBB9 Assessment of
biofilms formed by K. pneumoniae MBB9 on glass surfaces Table 1: Effect of substratum surfaces and time on bacterial adhesion at 37 °C Bacterial viability results showed that under both
conditions bacteria in a biofilm were higher after 12 hours compared to those
with damaged membranes. Under shaking conditions, cells with intact membranes
were a few higher with the same order of magnitude; however, dead cell numbers
increased with the increase of incubation time from 12 to 24 to 48 hours
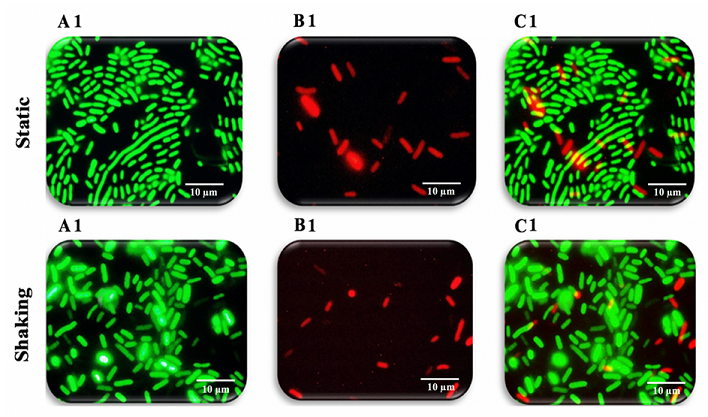
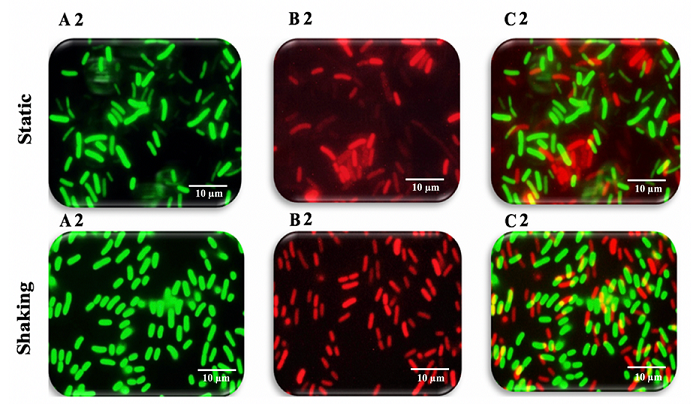
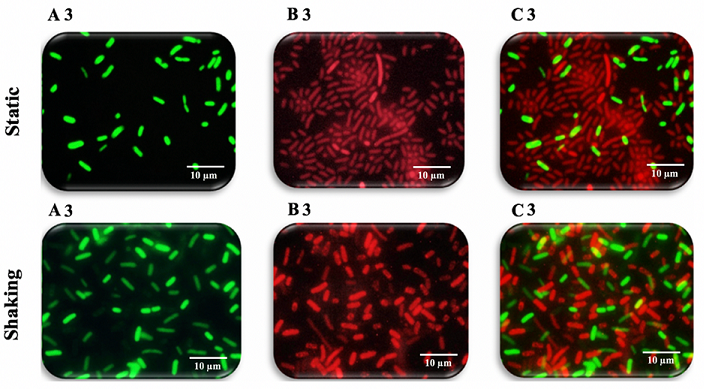
(Figures 9-11). Figure 9: Fluorescence microscopy images of K. pneumoniae MBB9 Figure 10: Fluorescence microscopy images of K. pneumoniae MBB9 biofilms Figure 11: Fluorescence microscopy images of K. pneumoniae MBB9 biofilms The capacity of biofilm formation of 10 gram-negative
potentially pathogenic bacteria showed that K. pneumoniae MBB9 had the greatest
biofilm capability to form biofilm in the microtiter
plate assay [35]. Therefore, K. pneumoniae MBB9 were chosen to assess the
impact of different factors on biofilm formation using the crystal violet and
resazurin assays. Biofilm formation is known to be powerfully affected by the
nutritional environment and the rate and amount of biofilm accumulation
increased with increasing carbon levels [19,36]. Biofilm in wells with 0.25 g
L-1 glucose showed high growth in comparison to the biofilm formed in the
remaining media, implying that 0.25 g L-1 glucose can be beneficial for K.
pneumoniae MBB9 biofilm (Figure 1). Ninety-three percent of Listeria
monocytogenes isolates, as previously demonstrated by Pan et al. (2010) [37],
formed higher biofilm in the presence of glucose than glucose-free medium.
Bühler et al. (1998) [17] have shown that glucose can enhance the quantity of
biofilm formed by E. coli and Burkholderia cepacia. For the same cultivation
conditions, the amount of biofilm significantly decreased with increasing glucose
concentrations in the medium to 0.5 g L-1, suggesting that glucose might be
useful for biofilm formation up to a certain limit. Hence, glucose metabolism might be a key factor in the
process of biofilm formation by K. pneumoniae MBB9, but increased
concentrations of glucose might lead cells to cell detachment from the biofilm.
Similarly, the density of biofilm by P. putida improved when increasing glucose
levels up to a limit (0.5 g L-1), whereas high glucose level (1 g L-1) was
found to decrease the rate of biofilm accumulation [19]. This was interpreted
as suggesting that biofilm formation can be regulated by the processes of detachment
in response to nutrient availability. The production of biofilm by E. coli was
found to raise linearly with rising glucose levels up to 10 mmol L-1, implying
that the total yield of biofilm could be controlled by the medium substrate
levels [17]. Jackson et al. (2002) [38] have shown that the addition of glucose
to different media might have an effect on biofilm formation in different
species of Enterobacteriaceae family, such as K. pneumoniae, Citrobacter
freundii, E. coli and Salmonella enterica due to the catabolite repression
system, implying that catabolite repression of the development of biofilm is a
prevalent theme in this family of bacteria. Carbon Catabolite Repression (CCR)
which is usually regulated by the second messenger cyclic AMP (cAMP) controls
glucose uptake, and thus the presence of glucose in the growth medium has been
proved to repress the production of cAMP in several bacteria [39]. In classical catabolite repression, glucose transport may
lead to dephosphorylation of enzyme IIAGlc of the bacterial phosphoenolpyruvate
(PEP): carbohydrate phosphotransferase system that restricts this protein from
stimulating membrane-bound adenylate cyclase (Cya); thus, leads to a reduction
in the concentration of the intracellular cAMP [38]. The decline in K.
pneumoniae MBB9 biofilm at 0.5 g L-1 glucose level might due to the active
detachment that requires enzymatic cleavage of matrix polymers or phenotypic
adaptation of the attached cells which is controlled by various physiological
mechanisms, such as quorum sensing and catabolic repression signals in
response to the availability of nutrients [19]. After dispersion, microbial
cells begin to accustom and form biofilm again, and the biofilm mass at 1 g L-1
glucose was comparable to that produced in the glucose-free medium, suggesting
that high glucose level has no impact on K. pneumoniae MBB9 biofilm. There is some information about the influence of glucose
concentrations on the production of biofilm, but little is known about the
effect of changing nitrogen levels, particularly peptone and yeast extract, in
the same process. Meat peptone, a protein from animal sources that broke down
into amino acids and peptides, is an important nitrogen source because its
nitrogen content exceeds 13% of its total content. Yeast extract is a multiple,
ill-defined blend of natural
sources and is known to be rich in nitrogen, amino acids, vitamins, and
carbon needed for the growth of microbes as its nitrogen content exceed 10% of
the total content [9]. The quantity of K. pneumoniae MBB9 biofilm was considerably
enhanced with increased concentrations of peptone and yeast extract, indicating
that higher levels of peptone and yeast extract might be useful for the
formation of K. pneumoniae MBB9 in microtiter plates (Figure 2 and 3). Peptone
has enhanced, as previously demonstrated, the development of biofilm by E.
coli, Lactobacillus rhamnosus and Mycobacterium avium [24,40,41]. In media with
a yeast extract, biofilm
thickness was higher than that in media with different peptone levels. This
might be due to the availability of nutrients as peptone consists only of
protein hydrolysis products, while yeast extract includes different kinds of
nutrients, such as trace metals and vitamins [42]. Klebsiella pneumoniae MBB9 biofilm mass tended to decline
considerably with rising pH (Figure 4). The highest biofilm formation was
observed at pH 4.5, indicating that the possibility of this microbe to form
biofilm under acidic conditions. It has been found that biofilms can form under
pH stress-induced conditions. Biofilm formation has been observed to develop
under pH stress-induced conditions [43].
The formation of biofilm can help bacteria to withstand
highly acidic environments as their gel-like compositions can lessen the
diffusion of ions and support the development of a pH gradient within the
extracellular matrix [44]. The growth at varying pH values might imply that
biofilm can also be affected by environmental conditions at the sites of
colonization. Nicolau et al. (2013) [44] have revealed that K. pneumoniae MBB9
isolates were able to form biofilm on various materials, such as glass,
polyester strip, and polystyrene under acidic environments pH 4.5 and neutral
pH 7. There was no significant difference between values at pH 5.5, 6.5, 7.5
and 8.5, whereas the biofilm mass by K. pneumoniae MBB9 under alkaline states
(pH 10.5) was considerably decreased (approximately 60% mass reduction). A previous study of 23 Staphylococcus epidermidis clinical
isolates showed that for most of the tested strains, the biofilm mass decreased
under highly alkaline conditions than those at pH 7 in polystyrene microtiter
plates [45]. Similarly, Zmantar et al. (2010) [43] have also demonstrated that
alkaline conditions have an inhibitory impact on S. aureus biofilm on 96-well
culture plates. This was interpreted as suggesting that the biofilm production
might depend on the pH content of the medium due to the effect on initial
bacterial attachment. In the same vein, Nostro et al. (2012) [46] have also
shown that alkaline solution might hinder bacterial colonization and hence
could be used to control the environmental risks associated with biofilm. The
notable inhibitory action of alkaline environments on the biofilm formation
observed here could suggest that bacterial cells failed to adhere to the
microtiter plate wells surface. It could also be due to the slower growth under
highly alkaline conditions. The media characteristics, such as pH may affect the
adherence by K. pneumoniae MBB9 as the physicochemical characteristics of the
surface of a bacterial cell can be modified by pH, and therefore the microbial
adhesion to the substrate [47]. In addition, alkaline pH could also impair the
normal development of biofilm that held at the microcolony
stage [46]. Also, the reduction in biofilm at alkaline pH might be due to
the weak electrostatic repulsion between the cells and the substrate as the
amount of adhering cells was found to be enhanced when the pH of the medium was
near to the isoelectric point of the substrate surface, thus the electrostatic
properties of polystyrene surfaces may be influenced by pH value that might
lead to a weaker interaction with surfaces and thereby compromise the bacterial
adhesion [43]. Temperature is one of the important factors that affect
bacterial growth and biofilm development, and the majority of pathogens are
mesophiles that grow well at optimum temperatures between 25 °C and 40 °C [18].
Results showed that K. pneumoniae MBB9 biofilm density was highest at 40 °C
(Figure 5). Similarly, K. pneumoniae MBB9 were shown to form more biofilm on
glass and polystyrene surfaces at 40 °C than at 35 °C [44]. It has also been
reported that Pseudomonas putida tolerate temperatures of 40 °C compared with
30 °C by enhanced the biofilm production, suggesting a complex multilevel
regulatory process in which many different genes
are involved [48]. Maximum production for biofilms at 40 °C might suggest that
40 °C is the ideal temperature for biofilm development (Figure 5). Bonaventura
et al. (2007) [49] have shown that Stenotrophomonas maltophilia have the
capability of forming biofilm at room temperature, 32 °C and 37 °C. This was
interpreted as suggesting that slower bacterial growth might lead to the lower
absorbance obtained at room temperature. Similarly, the decrease in K.
pneumoniae MBB9 biofilm at 32 °C compared to 37 °C was also interpreted as
being as a result of the slowly growing bacteria at 32 °C [50]. The reduction in biofilm by K. pneumoniae MBB9 at 25 °C
might be due to the slower growth of bacteria. More extreme temperatures (50
°C) might negatively affect the formation of biofilm by inhibiting growth. A
study of the temperature on the polymeric and mechanical properties of
Staphylococcus epidermidis
biofilm has shown that morphology, cell viability, and mechanical
characteristics of bacterial biofilm can be affected by higher temperatures
that can generate a remarkable decrease in the yield stress and the biofilm
small strain elastic modulus, suggesting that high temperature can reduce the
integrity of the biofilm as increasing temperature close to the bacterium
optimum growth temperature has been found to improve the bacterial biofilm
elasticity [8,51]. The biofilm formation under aerobic conditions has been well
investigated, but little is known about biofilm formation under anaerobic
environments. Under anaerobic conditions, Klebsiella pneumoniae MBB9 were able
to form biofilm. However, biofilm production was lower (83% less) compared to
that achieved under aerobic conditions. Similarly, Worlitzsch et al. (2002)
[52] noted that P. aeruginosa produced further biofilm when grown
anaerobically. Pseudomonas aeruginosa can enter into the adherent mucus and
grow in hypoxic/anaerobic slime, and the potential of these bacteria to
proliferate in this area would generate fully hypoxic (anaerobic) environments
as the enhanced formation of alginate may serve as a stress response to hypoxia
that is part of the process that forms biofilm like micro colonies. Similarly,
some E. coli clinical strains were able to yield biofilms under anaerobic
conditions, suggesting that the ability of biofilm
formation is strain-dependent [10].
The biofilm formation ability under anaerobic conditions
seen here may suggest that the facultative anaerobe K. pneumoniae MBB9 can
produce biofilm in anoxic environments using available alternative electron
acceptors. Moreover, facultative anaerobes' growth may play a role in the
formation of biofilm in the environment by anaerobes. For example, in dental
plaque biofilm formation, many strict anaerobes, such as Bacteroides forsythus
and Fusobacterium nucleatum need the former attachment of an aerobe or a
facultative microbe to start the biofilm formation [36]. Sodium
chloride has been tested as an osmotic agent to study its effect on
microbial biofilm formation and development [2,53]. Results showed that biofilm
formed by K. pneumoniae MBB9 decreased with increasing NaCl concentrations in
the medium from 0.6% to 10.6% with the highest biofilm occurring at 0.6%
(Figure 7). The biofilm decline might suggest that higher concentrations of
NaCl negatively affect the ability of K. pneumoniae MBB9 to produce biofilm
through an osmotic impact or might due to weak growth of cells under high
levels of NaCl. These findings are in accordance with an earlier study
revealing that raises in NaCl levels from 0.5 to 10.5% suppress the formation
of biofilm by Salmonella in 96-well polystyrene microtiter plates [2].
Similarly, a study by Rinaudi et al. (2006) [54] has shown that high osmotic
potential limits the biofilm establishment produced by Sinorhizobium meliloti,
suggesting that NaCl negatively affects the formation of biofilm by an osmotic
force. Consequently, the inhibitory influence of NaCl on biofilm formed by K.
pneumoniae MBB9 could be a result of an osmotic impact or could be due to a
particular ion effect [54]. The growth media structure has been reported to affect the
ability of bacteria to form biofilm in vitro [55]. LB and TSB broths were both
supported the biofilm development of K. pneumoniae MBB9; however, higher
biofilm was yielded significantly in NB (Figure 8). This indicates that
nutrient provision might influence the ability of biofilm formation by K.
pneumoniae MBB9. However, compared to the biofilm formed in the TSB medium, the
quantity of biofilm was higher in the LB medium. The reduction of biofilm could
presumably be a result of the glucose that present in the TSB medium (2.5 g L-1
(0.25%)) compared to the LB medium that lacks glucose. The concentration of 0.5 g L-1 glucose in this study was
found to reduce the biofilm produced by K. pneumoniae MBB9 on polystyrene
plates (Figure 1). Jackson et al. (2002) [38] have noted that glucose addition
to different culture media inhibited the biofilm formation in several species
of Enterobacteriaceae family, among them K. pneumoniae, due to catabolite
repression of the biofilm development. Besides, the medium that facilitates the
high production of microbial biofilm might vary for each microbe [56].
Stepanovic et al. (2004) [57] has been shown that Salmonella spp. formed more
further biofilm in nutrient‐poor media, while L. monocytogenes
were observed to produce more biofilm in nutrient‐rich media, suggesting that
the structure of medium nutrients influenced the biofilm mass in various ways.
In the same vein, a study by Hood and Zottola (1997) [58] on five different
bacterial isolates has shown that the medium allowing a high level of adherent
cells can be varied for each microbe. So from the results, it can be concluded
that K. pneumoniae MBB9 biofilm can be cultivated in LB and TSB broths, but NB
medium might be more suitable since more biofilm was yielded in such
medium. Crystal
violet provides a good biofilm mass measurement but does not estimate the
biofilm viability as it quantifies the matrix of biofilm, including viable and
dead bacterial cells [59,60]. Accordingly, the resazurin assay is used to
determine the active biofilm cells as it only quantifies the viable bacterial
cells within the formed biofilm [60]. An increase in metabolic activity leads
to increase the resazurin conversion (oxidized) to fluorescent resorufin
(reduced) since a higher rate of resazurin uptake by bacterial cells could
cause a higher fluorescence reading [61]. Although the resazurin assay showed
the activity of K. pneumoniae MBB9 biofilm cells, some of the obtained crystal
violet assay results were not in accordance with those from the resazurin assay
(Figures S1-S8 in supplementary material). The culture medium has presumably
influenced the values of fluorescence and thus resazurin assay outcome [62,63].
However, decreased pH can lead to a reduction in the fluorescence intensity of
the resazurin [64]. From the results, highly acidic conditions had no effect on
the viable bacterial cell numbers biofilm; however, the trend was comparable to
that obtained using the crystal violet assay (Figure S4 in supplementary
material). In vitro, biofilms formation by K. pneumoniae MBB9 was
evaluated using the glass coupons incubated under both static and shaking
conditions at 37 °C for 12, 24, and 48 hours. The number of cells attached to
surfaces or associated with biofilms was quantified using the plate count
technique. Microbial adhesion to surfaces is the first step in colonization,
invasion, and biofilm formation [65,66]. Most bacteria have the ability to form
biofilms on abiotic and biotic surfaces [67]. The attachment of biofilms can
occur readily on surfaces that are rough, hydrophobic,
non-polar and coated by surface conditioning films [66]. The presence of
extracellular appendages, including flagella, pili, fimbriae and
exopolysaccharide, and the adhesion properties of surfaces, such as roughness,
are the most important factors that can affect the ability of cells to adhere
to the substratum [68]. Since the ability of microbes to form biofilms may
differ depending on the substrate, the adherence by K. pneumoniae MBB9 to
abiotic surfaces, such as hydrophilic glass was tested, and the bacterial
adhesion was expressed as CFU per cm2. It is generally accepted that hydrophobic materials can
provide a greater bacterial adherence [69,70]. It has been found that Listeria
monocytogenes can adhere in higher numbers to materials that are more
hydrophobic [71]. Karunasagar et al. (1996) [72] have been reported that the
variation in the density of biofilms depends on the surface's properties.
Results revealed that K. pneumoniae MBB9 possesses a high capacity to form
biofilms on the surface of glass slide coupons. Similarly, it has been reported
that K. pneumoniae species isolated from rivers and drinking water distribution
systems can adhere to glass, plastic (polycarbonate,
chlorinated polyvinyl chloride), and carbon steel [44,73,74]. Microbial load of
biofilms by K. pneumoniae MBB9 after 12 hours of incubation under static
conditions was statistically more compared with shaking conditions with values
of 6.60 × 104 and 5.50 × 104 CFU per cm2, respectively (Table 1). In this study, 12 h
incubation was found to be most efficient in biofilms formation by K.
pneumoniae MBB9 on the glass surface under both conditions. The duration of the
incubation period can considerably influence the production of biofilms as the
amount of biofilms increases with the incubation period [11]. Although the
density of biofilms was higher during the initial phase, the amount of biofilms
formed tended to decrease significantly (p ˂0.05) for both conditions over
time. Significant differences (p ˂0.05) were found between the biofilms formed
by K. pneumonia MBB9 under both conditions between the control and the
different incubation periods and between each incubation time. It is possible
that some biofilm associated cells composing the outermost layers of the
biofilm changed to the free-floating state, while others may have died as a
consequence of nutrient depletion and exposure to accumulated toxic metabolic
wastes generated by the biofilm after 24 hours of incubation. The incubation
period can be defined as the contact time between the surface and the bacterial
cells [75]. More biofilms were obtained during shaking conditions
compared with static conditions for the 24 and 48 hours of incubation with
values of 3.00 × 104, 2.45 × 104, 1.96 × 104 and 1.20 × 104 CFU per cm2,
respectively. Significant differences (p ˂0.05) were found between the biofilms
formed by K. pneumoniae MBB9 under static and shaking conditions for 12, 24 and
48 hours. It is evident that shaking conditions appear to enhance the cell
adhesion to the glass surface as a better degree of mixing can provide higher
nutrient levels to K. pneumoniae MBB9 cells, increase the oxygen
transfer rates, and thus promoted the bacterial growth within the biofilm.
It is known that shear forces may have an impact on the structure, production
of exopolysaccharide, mass transfer, and metabolic/genetic behaviors of
biofilms [76]. Zhang et al. (2011) [77] have been stated that high hydrodynamic
shear may lead to the detachment of Pseudomonas aeruginosa from the surface. The
multiple effects of hydrodynamic and nutrients were probably enhanced the
bacterial growth as there were no shear forces promoting cell detachment under
these low shaking conditions. It has been stated that shear stress is a
dominant factor that can influence the biofilm formation strongly when the
fluid flows at a higher velocity over the biofilm surface [76,78]. Some
materials, such as glass, stainless steel, and mica are hydrophilic and
negatively charged, while the plastic surface is more suitable for bacterial
attachment due to its hydrophobic nonpolar nature with little or no surface
charge [69,71]. To obtain a more definite result, a Live/Dead BacLight
Bacterial Viability Kit was used to monitor the viability of bacterial
populations. This kit is widely used for the enumeration of bacteria [79,80].
It consists of two nucleic
acid stains: SYTO9, which penetrates cell membranes more easily, and
propidium iodide, which is a highly charged stain that cannot permeate cells
but can penetrate damaged membranes [81]. Application of both dyes results in
intact cell membranes stain fluorescent green, whereas dead cells, because of a
compromised membrane, show intense red fluorescence [82]. Biofilms on each
glass chips were stained and incubated at room temperature for 15 minutes
before fluorescence microscopy examination. Images of the single focal plane or
vertical section of the stained biofilms were captured using a fluorescence
microscopy system. As can be seen in
Figure 9A1 and 9B1, the ratio between living and dead cells. Although the
viable bacteria in a biofilm were higher compared to those with damaged
membranes after 12 hours under both conditions, cells with intact membranes
under shaking conditions were a little higher compared to their counterparts
under static conditions with the same order of magnitude. The number of dead
cells increased with the extension of incubation time from 12 to 24 to 48 hours
as a result of the depletion of nutrients and exposure to metabolic byproducts
(Figures 9B1 static, 9B1 shaking, 10B2 static, 10B2 shaking, 11B3 static and
11B3 shaking). It is concluded that K. pneumoniae MBB9 strains study are
capable of adhering, colonizing, and forming biofilms on glass surfaces. The effect of environmental conditions on bacterial
behaviour has been extensively studied on planktonic cells, but more
information about how these factors modulate biofilm formation is required. In
the present investigation, some conclusions can be drawn: 1.
The biofilm in 0.25 g L-1 of glucose exhibited
greater growth in comparison to the biofilm formed in the remaining media. 2. The K. pneumoniae MBB9 biofilm profiles for the
different peptone and yeast extract concentrations were similar where the
biofilm increased with increasing the level of peptone and yeast extract and
the highest absorbance values were obtained for peptone and yeast extract concentration of 1 g L-1. 3.
The peak of K. pneumoniae MBB9 biofilm was at pH
4.5 and a significant decrease in the absorbance values occurred with further
pH increasing until (pH 10.5). 4.
Klebsiella pneumoniae MBB9 biofilm increased up
to 40 °C before decreasing with more extreme temperatures (50 °C). 5.
The amount of biofilm was maximal in the control
media and tended to decrease with increasing NaCl concentrations until (10.6%). 6.
Under anaerobic condition, K. pneumoniae MBB9
biofilm was lower (83% less) than that obtained under aerobic conditions. 7.
Klebsiella pneumoniae MBB9 have the capacity to
form biofilms on the surface of glass slide coupons. 8.
The amount of biofilms at 12 hours of incubation
under static conditions was statistically higher compared with shaking
conditions with values of 6.60 × 104 and 5.50 × 104 CFU per cm2, respectively. 9.
The number of biofilms formed under both static
and shaking conditions tended to significantly decrease over time.
Special thanks to the entire group in the Department of
Molecular Biology and Biotechnology at the University of Sheffield and in
particular Prof. Milton Wainwright and Prof. Jeff Green for their assistance. Ghazay F Alotaibi, Department of Environment and
Marine Biology, Saline Water Desalination Technologies Research Institute, P.O.
8328 Al-Jubail 31951 Al-Jubail, Saudi Arabia, E-mail: DAlotaibi@swcc.gov.sa Alotaibi GF and Bukhari MA. Characterization
and evaluation of biofilm formation by Klebsiella
pneumonia MBB9 isolated from epilithic biofilms of the porter brook river, Sheffield
(2021) Edel J Biomed Res Rev 3: 14-24 Biofilm Formation, Klebsiella pneumoniae, Microtiter Plate, Quorum Sensing, Environmental Factors, Alamar Blue, ResazurinCharacterization and Evaluation of Biofilm Formation by Klebsiella pneumonia MBB9 Isolated from Epilithic Biofilms of the Porter Brook River, Sheffield
Abstract
Full-Text
Introduction
Material and
Methods
The same day (March 2015), river-stones (thick, light brown, sticky growth) on
the upper surfaces were collected in a sterile plastic
container from Porter Brook in Sheffield, United Kingdom, and were stored
until analysis in the cool icebox.
Epilithic biofilms on the stones were scraped and suspensions were serially
diluted 1:100 in physiological saline (0.85%) using an aseptic technique [21].
Different selective media: R2A agar, Eosin-Methylene Blue (EMB) agar, MacConkey
agar, Xylose
Lysine Deoxycholate (XLD) agar, nutrient agar, and Violet Red Bile (VRB)
agar were used to inoculate the suspension. The inoculated plates were
incubated aerobically at 37ºC for 24-72 h. To obtain pure cultures of the
bacterial isolates, colonies with various colors and morphologies were streaked
again on freshly agar plates.
Isolated colonies from the agar plates were selected and characterized for
preliminary identification using various morphological and biochemical
properties [22]. Morphological parameters, such as colony form, elevation,
margin, surface, optical features, consistency and color are used along with
biochemical tests, including catalase and oxidase activities.
GenElute™ Bacterial Genomic DNA Kit was used according to the manufacturer's
instructions to extract bacterial genomic DNA from all isolated bacteria. DNA
preparations purity was assessed spectrophotometrically using a Nano drop 1000
(A260/280) (Nano
Drop Technologies, Wilmington, DE, USA). PCR was used to amplify 16S rRNA
genes of the bacterial isolates using forward (27F) 5’AGAGTTTGATCCTGGCTCAG-3’
and reverse (1492R) 5’GGTTACCTTGTTACGACTT-3’ primers (universal, 16S rDNA gene)
to amplify the V1-V9 region (1500 bp) of the16S rRNA gene. In brief, a master
mix of 25 µl total volume was prepared as follows: 12 µl of 2X master mix
(BioLabs, England), 2 µl of each oligonucleotide primer (10 µM), 7 µl of
Molecular Grade Water and 2 µl of template DNA. All reactions were run on a LabCycler
(SensQuest, Germany) under the following conditions: initial denaturation at 98
°C for 30 s, 35 cycles of 95 °C for 1 min, 58 °C for 30 s, 72 °C for 5 min and
a final extension at 72 °C for 5 min followed by a hold at 4 °C. The amplified
DNA fragments were separated on a 1% (w/v) agarose gel electrophoresis
(BIO-RAD, USA).
The microtiter plate method of O’Toole and Kolter (1998) [23] was applied with
a few adjustments. Briefly, bacterial isolates were grown overnight in nutrient
broth at 37 °C. The OD600 of the bacterial suspensions
was adjusted to 0.5 McFarland standards (approximately 108 CFU/ml). A
flat-bottomed polystyrene 96-well microtiter plate (Costar; Corning
Incorporated., USA) was used to inoculate aliquots (200 μl). As plate sterility
controls, the sterile nutrient broth was used and plates were covered and
incubated at 37 °C for 24 h [24].
The metabolic activity of adherent cells was determined using resazurin
(7-hydroxy-3H-phenoxazine-3-on-10-oxide). The microtiter plate method of
O’Toole and Kolter (1998) was applied with a few adjustments. Briefly,
Klebsiella pneumoniae MBB9 was grown overnight in nutrient broth at 37 °C. The
optical density of the bacterial
suspensions at 600 nm was adjusted to 0.5, equivalent to 108 CFUml-1 [27].
Aliquots (200 μl) were then inoculated into wells of a flatbottomed polystyrene
96-well microtiter plate (Costar; Corning Incorporated., USA). Plates were
covered and incubated at 37 °C for 24h. Following the incubation, cells in
planktonic forms in the fluid were discharged by inverting the plate and
thoroughly rinsed three times with 200 μl of sterile deionized water (dH2O) to
remove unattached bacteria. The wells were then loaded with 180 μl of sterile
nutrient broth and 20 μl of resazurin solution (Invitrogen™). The plates were
immediately covered with aluminium foil and incubated at 37 °C in the dark for
4 hours according to the manufacturer’s instructions. Using the microplate
reader (BioTek FLx800, UK), fluorescence signals (λexcitation: 540 nm and
λemission: 580 nm) were measured. All experiments performed in triplicates and
the values of fluorescence were corrected by subtracting the readings from the
control.
Various levels of glucose, peptone and yeast
extract were individually changed to assess the impact of carbon and nitrogen
sources on biofilm formation by K. pneumoniae MBB9 in microplates using crystal
violet and resazurin assays.
Effect of pH on biofilm formation: Nutrient
broth medium adjusted to various pH values 4.5, 5.5, 6.5, 7.5, 8.5 and 10.5 was
used to test the impact of pH on biofilm formation by K. pneumoniae MBB9 in
microplates using crystal violet and resazurin assays.
Biofilm formation on glass coupons:
The method of Adetunji and Isola (2011) [28] was used with a few alterations to
investigate the impact of substratum type on biofilms formation by Klebsiella
pneumoniae MBB9. Briefly, an overnight bacterial culture was grown under
aerobic conditions to the mid-log phase in nutrient
broth at 37 °C with shaking. The OD600 of the bacterial suspensions was then
adjusted to 0.5 McFarland standards (approximately 108 CFU/ml) in a fresh
nutrient broth medium. The glass slide coupons (2.5 cm × 2 cm) were soaked in
detergent, washed with sterile deionized water (dH2O) and dried by air before
being autoclaved at 121 °C for 15 min. Every plastic beaker holding sterilized
glass coupon immersed in 9,000 µl sterilized nutrient broth was then inoculated
with 1,000 µl of the bacterial suspension [29]. As sterility control, the
sterile nutrient broth was used, and then containers were incubated under
static (without agitation) and shaking conditions with a low shear fluid for
12, 24, and 48 hours.
The results were used to calculate the mean and Standard Deviations (SD). In
the analysis of data, one-way ANOVA was used. Statistically significant p
values < 0.05 were considered. The error bars represent the default
deviation. Results
As shown in Figure 1, the maximum amount of biofilm was at 0.25 g L-1 glucose
and then the number of biofilm significantly (p ˂0.05) diminished with
increasing the levels in the medium to 0.5 g L-1 and remained relatively
consistent for up to 1 g L-1 glucose. The biofilm mass significantly (p ˂0.05)
increased with increasing the concentrations
of peptone and yeast extract (Figure 2 and Figure 3). 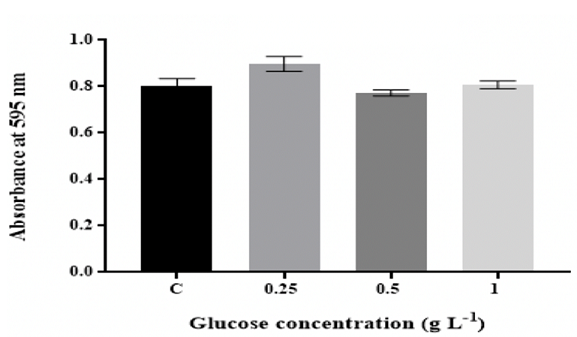
The findings showed that K. pneumoniae MBB9 were capable to form biofilm under
acidic pH conditions and the quantity of biofilm reduced significantly (p
<0.05) at higher pH values (Figure 4).
The production of biofilm was maximal at 40 °C, followed by 37 °C, whereas the
biofilm thickness was significantly (p ˂0.05) reduced at 25 °C and 50 °C
(Figure 5). 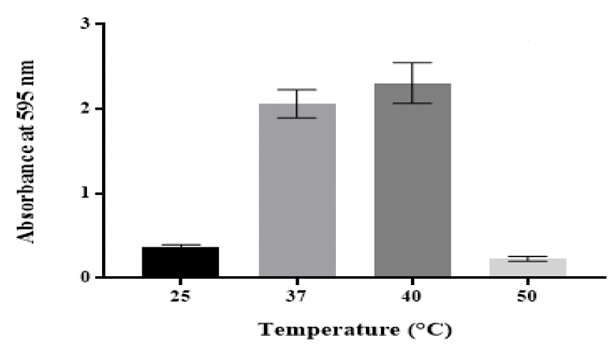
From the results, K. pneumoniae MBB9 have an ability to form biofilm under
anaerobic conditions, albeit less than under aerobic conditions (Figure
6). 
The findings showed that the number of biofilm by K. pneumoniae MBB9 decreased
significantly (p ˂0.05) with increasing levels of NaCl, with maximum biofilm
formation being seen in the unadjusted nutrient
broth (C: 0.6% NaCl) (Figure 7).
From the results, the quantity of biofilm formed in NB was considerably (p
˂0.05) higher compared to other media, approaching a maximum in NB followed by
LB and subsequently by TSB (Figure 8). 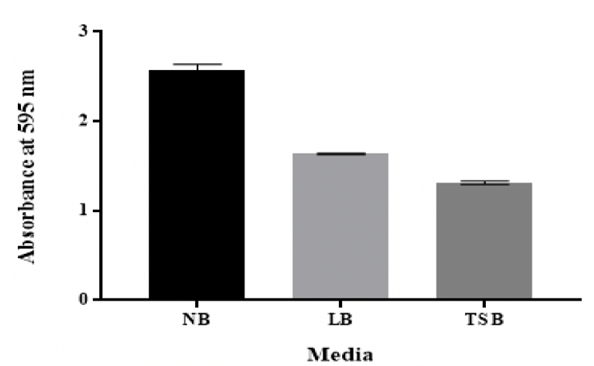
The findings showed that K. pneumoniae MBB9 were capable to form biofilm on the
glass slide coupons surfaces. The number of biofilm by K. pneumoniae MBB9 after
12 hours of incubation under static conditions was statistically (p ˂0.05) more
compared with shaking conditions with values of 6.60 × 104 and 5.50 × 104 CFU
per cm2, respectively (Table 1). In both conditions, the quantity
of biofilms formed tended to decrease significantly (p ˂0.05) over time (Table
1). 
Discussion
Conclusion
Acknowledgements
References
Citation
Keywords