Research Article :
During the summer 2014, dark black symptoms
beneath the leaf surfaces were observed on the L species in the south Kashmir. In the later stages of
development the infection subtends the whole surface resulting in leaf
collapse. Fungal isolates were recovered directly from the structures present
on the lesions. Purified DNA from each isolate was amplified using the random
amplified polymorphic DNA technique recruiting the ITSI (TCCGTAGGTGAACCTGCGG)
and ITS4 (TCCTCCGC TTATTGATATGC) specific primers. Amplification products
visualized on agarose gel showed specific band pattern for each isolate. The mycotaxonomic
characterization and phylogenetic interpretations showed the emergence of
fungal species, Meliola sp. KY623717 with genetic variance of 2% in internal
transcribed spacer (ITS) region of rDNA from closely related species under
geographically distinct region. Black colony forming
parasitic fungi are
known as Black or dark mildews. The Meliola species are obligate bio trophic
parasites found superficially on the aerial portions of vascular plants
commonly known as black or dark mildews [1]. These are ecto-parasites producing
black colonies on the surface of the host plants. The black colony forming
parasitic fungi belong to several taxonomic groups, viz. Hyphomycetes,
Meliolales, Schiffnerula and its anamorphic forms, Asterinales, Meliolinaceae,
etc. Of these, the fungi belonging to Meliolales can be distinguished by their
two celled appressoriate mycelium,
setae, presence of globose perithecia with setae, appendages, etc. These are
the unique group of fungi which are distinguished very easily. This work gives
a clue to the identification of the meliolaceous fungi at a glance based on the
morphological as well as molecular data. DNA Isolation Dry mycelium mat (50
mg) was transferred in a sterile eppendorf tube. It was crushed with sterile
scissor and made fine powder. CTAB extraction buffer (600 µl) pre warmed at 60 °C was also added [2].
The resulted mycelial powder was incubated in a water bath at 50 °C for 3–4 h with
vortexing at the interval of 10 min. The resulting samples were centrifuged at
10,000 rpm for 20 min at room temperature. The supernatant was transferred to
sterile eppendorf tube. Chloroform (600 µl) and isoamyl alcohol (in 24:1
ratio) were added and subjected to vortexing again for 5 min. Sodium acetate
(0.5 µl, 3 M, pH 5.2) was added further and mixed gently and stored in ice for
15 min. The solution was again
centrifuged at 16,000 rpm at 4°C
for 10 min. Chilled isopropanol
(600 µl) was added to eppendorf tube and stored at -50°C for 10 min. The supernatant
was decanted and 600 µl ethanol (70%) was added and solution was kept at room
temperature. The overnight pellet was dried. Distilled water (25 µl) was added
and kept for 2 hr at room temperatures followed by electrophoresis. The DNA
samples were stored at -20 °C. ITS Amplification
and Gene Sequencing Universal
primers ITSI (TCCGTAGGTGAACCTGCGG) and ITS4 (TCCTCCGC TTATTGATATGC) [3] were
used to amplify the ITS regions between the small (18S gene) and large (28S
gene) nuclear rDNA, including the 5.8S rDNA. Amplifications
were carried out in a 50 µl volume, containing 10 mM Tris–HCl pH 8.3, 50 mM
KCl, 1.1 mM MgCl2 and 0.01% gelatin, 200 l M of each dNTP (Promega), 1.0 l M of
each primer, 1.0 U of Taq DNA polymerase (Sigma) and 30–50 ng of DNA. Amplicons
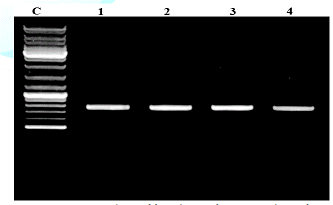
were visualised on 1.5% agarose gel under the UV trans-illuminator (Figure 1).
Cycling parameters were 5 min at 94°C; 30 cycles of 1
min at 94°C, 1 min 30 s at 50 °C,
1 min 30 s at 72° C; with a final extension at 72 °C
for 7 min. Aliquots of about 35 ng of DNA were purified from agarose gel with a
Qiaex II Kit (Qiagen, Hilden, Germany) and automatically sequenced with an ABI
Prism 3100 DNA Sequencer (Applied Biosystems, Norwalk, CT, USA). Phenotypic
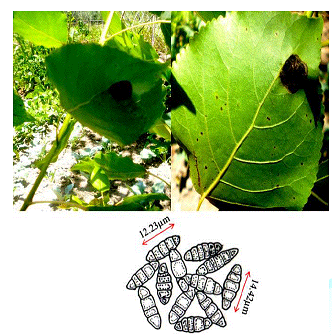
observations Dark
black symptoms on beneath the leaf surfaces were
observed on the Populas alba in the south Kashmir. In the later stages of
development the infection subtends the whole surface which results in the leaf
collapsing (Figure 2). Minute
amphigenous black colonies mostly epiphyllous, initially scattered but becoming
confluent with age, dense, 1–6 mm diameter. Hyphae dark brown, septate,
straight to sub-straight bearing appressoria. Perithecia brown, globose,
scattered, black, 102–127 μm diameter [4]. Ascospores hyaline inside the ascus,
becoming grey or brown with age, dark brown or grey at maturity, cylindrical to
ellipsoid, rounded at the tips, 4-septate, constricted at the septa. Genotypic
observations PCR
products analyzed on 2% agarose electrophoresis gel. Primers (5-ACCCGCTGAACTTAAGC-3) and (5-TCCTGAGGGAAACTTCG-3)
were used to amplify the partial ITS regions. For phylogenetic analysis, ITS
rDNA sequences from additional species were retrieved from GenBank. The
phylogenetic tree based on concatenated ITS regions of rDNA strict consensus
sequences of the Meliola mangiferae KY623717
and other fungal lineages used as out groups [5,6]. Consensus regions were
compared against GenBank database using Mega BLAST program. The closest hit
sequences and representatives of selected Meliola species were obtained from
GenBank (http://www.ncbi.nlm.nih.gov/) to help clarify the phylogenetic
relationship of Meliolales within the class. All sequences were downloaded in
FASTA format and aligned using the multiple sequence alignment program MUSCLE
built in PHYML (Phylogenetic Inferences using Maximum Likelihood). Alignments
were checked and necessary manual adjustments were made. All ambiguously
aligned regions within dataset were excluded from the analysis. Gaps
(insertions/deletions) were treated as missing data. Branches with < 30%
bootstrap support collapsed to polytomies and a long branch shortened by 50%.
Terminal nodes marked strict consensus sequences with the accession numbers
listed in supporting GenBank data base. The scale bar indicates the number of
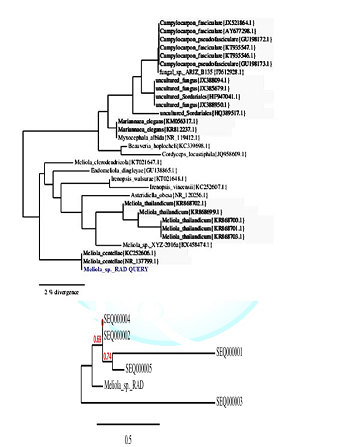
substitutions per site. In interpreting the tree it was found that Meliola mangiferae_ KY623717 has
diverged from closely related Meliola
centellae (KC252606.1) and Meliola centellae (NR_137799.1) by 2%. Phylogenetic
Analysis The
maximum likelihood tree was constructed using the MABL (Methodes et Algorithmes
pour la Bio informatique Lirmm) bioinformatic program to trace the phylogeny of
the collected Meliola species. The scale bar indicates the number of substitutions
per site. Meliola sp._ KY623717 has diverged from closely related Meliola centellae (KC252606.1) and Meliola centellae (NR_137799.1) by 2%
under geography distinct environment (Figure 3). The DNA sequence has been
deposited in NCBI GenBank with accession no KY623717. >Meliola mangiferae ACCTCGGATCAGGTAGGAATACCCGCTGAACTTAATCTCCGTTGGTGAACCAGCGGAGGGATCATTATAGAGTTAACAAAACAACTCCCAACCCTTTGTGAACCTTACCTACCGTTGCTTCGGCGGACCGCCCCGGGCGCTGCGTGCCCCGGACCCAAGGCGCCCGCCGGGGACCACACGAACCCTGTTTAACAAACATGTGTATCCTCTGAGCGAGCCGAAAGGCAACAAAACAAATCAAAACTTTCAACAACGGATCTCTTGGTTCTGGCATCGATGAAGAACGCAGCGAAATGCGATAAGTAATGTGAATTGCAGAATTCAGTGAATCATCGAATCTTTGAACGCACATTGCGCCCGCCAGTATTCTGGCGGGCATGCCTGTCCGAGCGTCATTTCAACCCTCGGGCCCCCCCCTTTTCCCCTCGCGGGGGAGGGGGCGGGCCCGGCGTTGGGGCCCAGGCGTCCTCCAAGGGCGCCTGTCCCCGAAACCCAGTGGCGGCCTCGCCGCTGCCTCCTCCGCGTAGTAGCACAAACCTCGCGGGCGGAAGGCGGCGCGGCCACGCCGTAAAACCCCAAACTTTTACCAAGGTTGACCTCGGAT The authors thank Head,
Department of Botany, Dr. Harisingh Gour Central University, Sagar, for
providing necessary laboratory facilities. They acknowledge Paul Hebert DNA
Centre, Aurangabad, for DNA barcoding and genetic analysis. Conflict
of interest: The authors declare that no conflicts
of interest exist for publication of this manuscript. 1.
Carlier G, Lorand JP, Bonhomme M
and Carlotto V. A reappraisal of the Cenozoic inner arc magmatism in southern
Peru: consequences for the evolution of the Central Andes for the past 50 ma
(1996) Third ISAG 551-554 2.
Schoch CL, Seifert KA, Huhndorf
S, Robert V, Spouge JL, et al. Nuclear ribosomal internal transcribed spacer
(ITS) region as a universal DNA barcode marker for Fungi (2012) Proc Natl Acad
Sci 109: 6241-6246. https://doi.org/10.1073/pnas.1117018109 3.
White TJ, Bruns T,
Lee S and Taylor J. Amplification and direct sequencing of fungal ribosomal RNA
genes for phylogenetics.
PCR protocols: a guide to methods and applications (1990) Academic Press,
SanDiego 315-322. 4.Crous PW, Lennox CL and Sutton
BC. Selenophoma eucalypti and Stigmina robbenensis sp. nov. from eucalyptus
leaves on Robben Island (1995) Mycol Res 99: 648-652.https://doi.org/10.1016/S0953-7562(09)80521-2 5.
Rafiq AD, Akhila NR and Imtiyaz
AS. First report of white stain of apple caused by Trichothecium kashmeriana in
India (2016) Arch Phytopathol Plant Pro 50. https://doi.org/10.1080/03235408.2016.1264178 6.
Rafiq AD, Akhila NR and Imtiyaz
AS. Stigmina carpophila detected on Prunus armeniaca and Prunus persica in
India (2017) Australas Plant Dis Notes 12:19. https://doi.org/10.1007/s13314-017-0245-6 Populas Alba, Leaf collapsing; Primers, Mycotaxonomic, Phylogenetic, Genetic variance
Implementation of Sanger DNA Sequencing Technologies in Tracing the Phylogeny of Meliola mangiferae
Rafiq Ahmad Dar, Akhila Nand Rai
Abstract
Full-Text
Introduction
Material
and Methods
Results
Acknowledgements
Compliance
with Ethical Standards
References
Keywords