Research Article :
Lan-Rong Chen, Jean Alain Trejaut,
Ying-Hui Lai, Zong-Sian Chen, Jin-Yuan Huang, Marie Lin and Jun-Hun
Loo Abstract The
genetic profile of Negritos of the Philippines differs from the non-Negrito
groups with mitochondrial DNA haplogroups B4b1a2, B5, D6a, M, M52a, and N11b.
Although Negritos are not seen in Taiwan, the strong genetic affinity between
the Philippines and Taiwan Mountain Tribe Aborigines (TwMtA), and Folks tales
of TwMtA, Saisiyat and Atayal recounting past contacts with Negritos, warrant
the search for a Negrito signature in Taiwan. Material and Method: Discriminant Analysis of Principal Component (DAPC)
was used to determine the genetic relationship between TwMtA, Filipino and
non-TwMtA groups. Results: The deep coalescence of B4b1a2 in the Philippine
Negritos, Saisiyat, Atayal, Island Southeast Asia, and SEA (Southeast Asia)
suggested a deeply rooted common ancestry, but could not support a past Negrito
presence in Taiwan. Conversely, the sharing of cultural components and mtDNA (mitochondrial DNA) haplogroup D6a2 in Saisiyat, Atayal and Philippine
Negritos may characterize a Negrito signature in Taiwan. Although the molecular
variation of D6a2 determines its presence in Taiwan back to middle Neolithic,
other markers, Y-SNP haplogroups C-M146 and K-M9, warrant further analysis. Most likely, the
physical characteristics, languages, and the genetic makeup of the Negritos in
Taiwan have been diluted as the result of heavy migration from the mainland in
the last 400 years. The Saisiyat tribe is an
Austronesian speaking group, a member of the Taiwan Mountain Tribe
Aborigines. In the year 2018 April, the Saisiyat
numbered 6,607, making them one of the third smallest indigenous groups on the
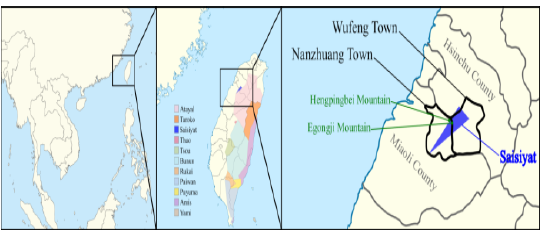
island [1]. Its people live in the North West flank of the Taiwan mountain
range, between Hsinchu and Miaoli (Figure
1). The geography of this region, which comprises the Egongji and
Hengpingbei Mountains, forced Saisiyat to divide into two groups, the
Sai-Kirapa in the north and the Sai-Maghahyobun in the south. Historically Sai-Kirapa has had
significant interaction with the northeastern Atayal tribe while the
Sai-Maghahyobun has had more contacts with the Hakka who migrated there from
East China in the last 400 years [2-6]. Similarly, Saisyat has two main
dialects: the Taai Dialect in the North, and the Tungho Dialect in the south [7].
While Saisiyat has traditional views that mix aspects of ancestor worship and animism
where all things are considered being alive and possess a distinct character,
other Saisiyat people also practice Christianity [2]. Figure
1: Saisiyat distribution map. Most TwMtA tribes have kept
folktales and myths that relate to past contacts with Negritos. In particular,
Saisiyat is the only tribe in Taiwan that has rituals every two years honoring
the memory of the Short People or Pas-ta ai [8]. The Short People in these
folktales are described as short-statured, dark-skinned and frizzy-haired and
have the same anthropometric characteristics as Negritos in the Philippines.
Some anthropologists believe these may have been Proto-Australoid people who
possibly arrived from Africa during the early Southern Dispersal 60,000 years
ago, but to this day, no archeological evidence has ever revealed the past
presence of Negritos in Taiwan [9,10]. It now proposed that, instead of a
shared ancestral phenotype from an ancient and well-distributed population, the
resemblance of Negrito with other Negritos of Asia and Pygmies in Africa is the
result of convergent evolution in the different parts of the word under
equivalent environmental conditions [11,12]. This is supported by genetic
evidence showing that Negritos of different parts of the world region have
different genetic structures [13]. Further, other genetic studies observed that
there is no simple dichotomy between Negrito and non-Negrito groups of the
Philippines [14,15]. Most Negrito groups share genetic
variations with neighbor populations while they have more deeply rooted
variants that suggest a much earlier arrival in the region, isolation, and
admixture with later arrivals [16]. Many studies using mitochondrial
DNA (mtDNA) and Y-chromosome variation have
established a significant common ancestry between the populations of Taiwan and
the Philippines, it is therefore expected to find a Negrito signature in Taiwan[14,15,17-21].
Although Atayal and Saisiyat have a genetic profile that distinguishes them
from the southern TwMtA, the polymorphism is homogenously distributed through
all the tribes. [2,3,6,21-24].In this study, we analyze the mitochondrial
genetic polymorphism of the Saisiyat tribe and search genetic evidence of the
speculated presence of Negritos in all Taiwan indigenous groups. The mitochondrial molecular clock
is faster than the molecular clock of Non-Recombination
Y-chromosome (NRY) haplogroups determined using
Single Nucleotide Polymorphisms (SNPs) and slower for NRY haplotypes determined
using Short Tandem Repeats (STRs) [25]. The rate of mtDNA is, therefore, most
appropriate to measure and trace evolutionary human changes phylogenetically in
time and space. Further, its short length (16,569 base pairs), its presence in
both males and females, its high polymorphism and the higher concentration than
genomic DNA, makes it a most effective material, practically and financially,
to use in a small laboratory [26]. However, we will use NRY in our comparative
analysis with the Philippines. Mitochondrial
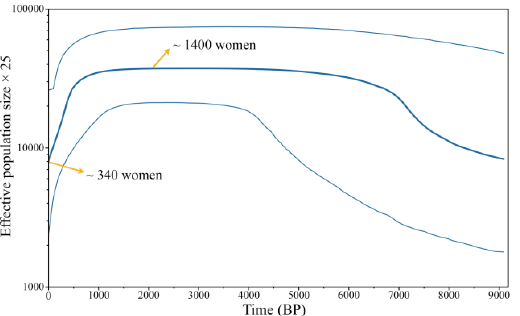
DNA Diversity Compared to the mtDNA genetic
diversity of all Taiwan groups (h=0.717 to 0.991), the Saisiyat tribe (h=0.864)
was lower than in Fujian and Taiwan Han (Minnan and Hakka, TwH) (~ 0.99), and
in range with other TwMtA (h=0.717 to 0.942) (Table 1) [27]. Similarly, the number of mtDNA
haplogroups observed in Saisiyat (n=20) was in
range with the number of haplogroups seen in other TwMtA (n=7 to 55) and
contrasted strongly with the number seen in Fujian, Minnan and Hakka (n=87, 95
and 226 respectively). Further, the tests of neutrality for Saisiyat, Tajima s
D (D=-0.438; p=0.365) and the more powerful Fu s Fs test (Fs=-24.275; p<0.0001)
indicated a departure from neutrality expectation, and were in range with most
value observed among other TwMtA groups (Data not shown). Demographic
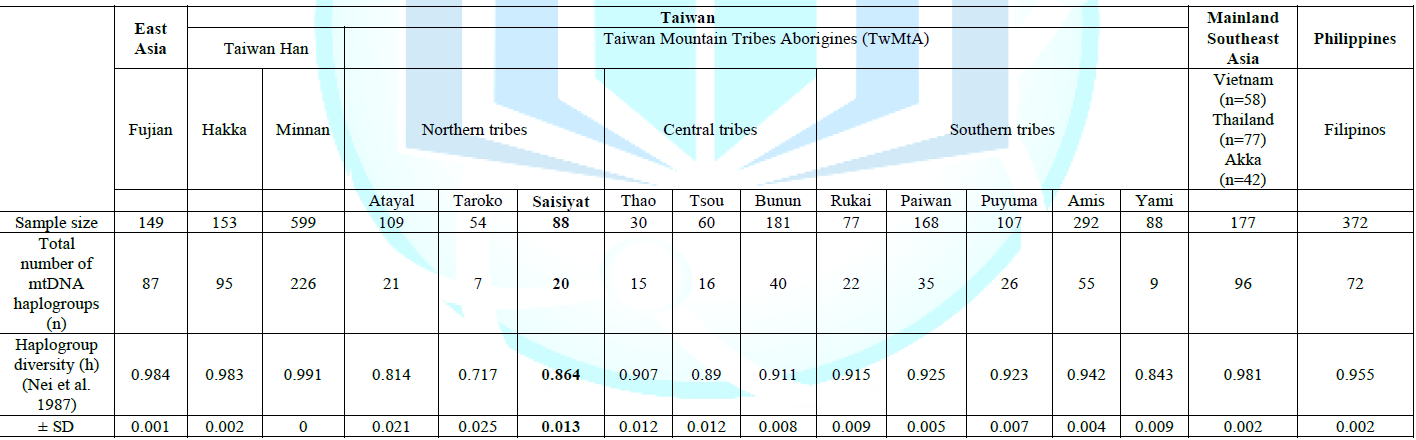
Analysis A bimodal curve was observed
between 11 and 21 basepair differences in the mismatch distribution analysis of
Saisiyat for 88 mtDNA sequences (Figure
2). The bimodal mismatch curve may have been the result of admixture.
Further, in contrast with the Fu s Fs test, the plot did not support population
sudden expansion [28]. Similarly, the hypothesis of sudden expansion was
rejected by two demographic indexes, the Sum of Squared
Deviation (SSD) test (SSD=0.008, P<0.001) and
the raggedness (r=0.011, P<0.001) (Harpending 1994) indicating that the data
deviated from the simulation expected under the model of expansion (Figure 2,
blue line). The analysis (Number of pairs vs. Base pair differences) was
obtained from 88 mtDNA Saisiyat sequences using nucleotide positions (nps)
8,000-9,000, nps 10,000-11,000, and Hyper
Variable Segment I (HVS-I) nps 16040-16390 [22,24].
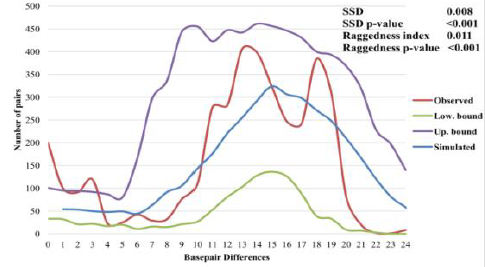
Using the Bayesian Skyline method, patterns of historical demography can also
be inferred from estimates of the effective population size over time. Figure 2:
Mismatch Distribution Analysis (MMDA) of Saisiyat. Accordingly, we constructed a Bayesian
Skyline Plot (BSP) plot (Figure 3) from 88 Saisiyat sequences of the mtDNA Hyper Variable
Segment I (HVS-I) data, with 20 million Markov Chain Monte Carlo (MCMC)
iterations, sampled every 3,000 steps, using a relaxed molecular clock and a
mutation rate of 2.2964 x 10-7 mutations per site per year. The BSP showed an
early Neolithic signature of population expansion, followed by a long phase of
relatively constant population size and a sudden steep population reduction
around 500 years BP. The Saisiyat present-day effective population obtained in
Figure 3 is approximately 340 women (CI 100 to 1120 women) [29]. Figure 3:
Bayesian Coalescent Skyline analysis of the Saisiyat
tribe. Bayesian Skyline Plot (BSP) plot
based on the mtDNA obtained from 88 Saisiyat sequences for the HVS-I control
region. BSP was calculated using a relaxed molecular clock and a mutation rate
of 2.2964 x 10-7 mutations per site per year and estimated with 20
million MCMC iterations sampled every 3,000 steps. The dark blue line represents
the posterior median of the effective population size through Time (one
generation=25 years). The light blue lines represent the 95% confidence
interval. MtDNA
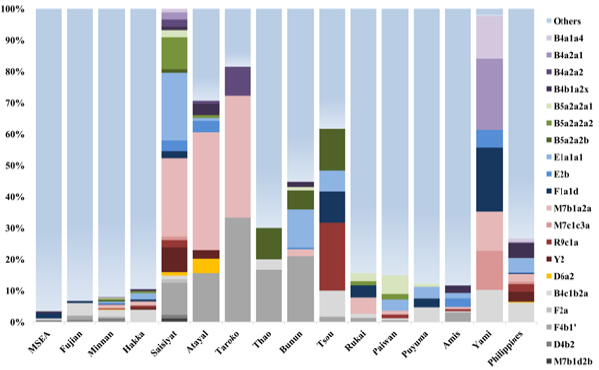
Haplogroup Distribution Out of 20 mitochondrial
haplogroups seen in Saisiyat (Figure 4),
15 were uniquely shared with the other the Austronesian speaking groups of
Taiwan, and five haplogroups (B5a2a2a2, E1a1a1, F4b1 , M7b1a2a and Y2) had a frequency
greater than 8% representing 75% of the Saisiyat gene pool. Interestingly,
haplogroups D6a2 was only seen in Saisiyat (1.1%), Atayal (4.6%), reported in a
single individual of the Pazeh plain tribe, and the Mamanwa Negritos from the
Philippines (3.3%) [14,24]. Figure
4: Haplogroup frequency in Saisiyat shared with other
populations. Distribution of shared mtDNA
haplogroups of Saisiyat constructed using Taiwan data and other relevant
populations [24]. Haplogroups with a shade of grey represent sharing with non-Indigenous
groups of Taiwan (Minnan and Hakka). All haplogroups not seen in Saisiyat but
present in other groups are represented by other . Population
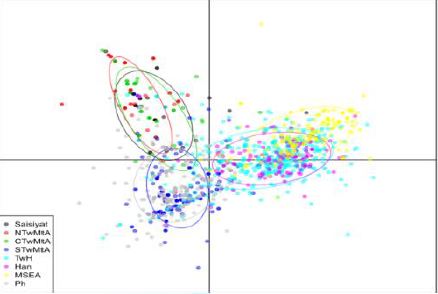
Differentiation Discriminant Analysis of
Principal Components (Figure 5)
showed a clear geographic divide along the first Discriminant component (the X
axis), which form a separates the Austronesian-speaking groups on the right,
from the Han, the TwH, and MSEA (Indochina) on the left. Further, although
individuals were not tightly grouped, the Northern and Central TwMtA form a
cluster that clearly separates them from a lower cluster encompassing Southern
TwMtA and the Philippines. Finally, we note that the Southern
Austronesian cluster (Southern TwMtA and the
Philippines) shows more admixtures with TwH than the Northern cluster. The dots represents individual
from eight different groups and locations whose inertia ellipses are
characterized by a color as indicated in the insert on the left. Sampling
locations are: NTwMtA: North Taiwan Mountain Tribe Aborigines (TwMtA); CTwMtA:
Central TwMtA; STwMtA: Southern TwMtA; TwH: Taiwan Han (Minnan and Hakka);
MSEA: Mainland Southeast Asia (Indochina); Ph: Philippines; Han: Fujian. Phylogenetic
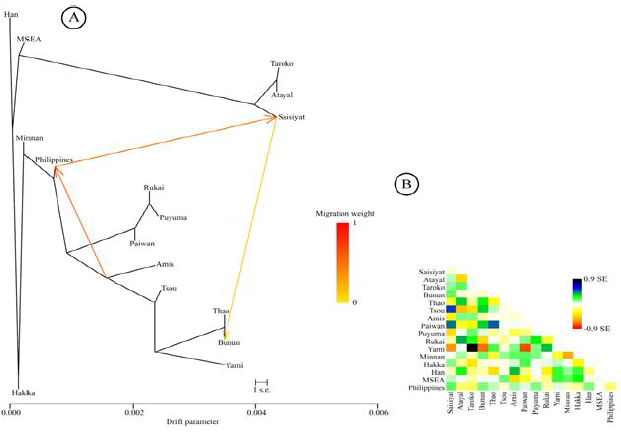
Analysis A maximum-likelihood tree using
mtDNA haplogroup frequencies was inferred with the TreeMix software (Figure 6A).
The tree was consistent with the geographical distribution of populations in
Taiwan; it places Saisiyat in a strong relationship with the Northern tribes
(Atayal and Taroko). Further, migration arrows were first limited to 20 migration
events in the analysis and only the three most significant gene flow events
were retained for clarity of (Figure 6B). The relationship delineated between
Saisiyat and the central TwMtA (Thao and Bunun) was expected from the DAPC
clustering shown in Figure 5. More interesting, was a significant input from
the Philippines to Taiwan, consistent with a previous study [20]. And here seen
as gene flow to Saisiyat with a migration weight of ~ 0.65 (Figure 6). Most likely, this result is
the effect of the sharing of haplogroups B4a2a, E2b, M7b1a2a, D6a2, R9c1a and
Y2 between Saisiyat and the Philippines (Figure 4). 6A: Maximum-Likelihood tree
inferred by TreeMix for all Taiwan and SEA populations assuming 20 significant
gene-flow events, only the three most significant gene-flow events are shown
for clarity, they are colored according to their weight on a zero to one scale
[30]. 6B: Residual fit from A. Shaded colored cells represent Standard error
for admixture events across all pairs of populations. Population pairs with a
residual above zero are more closely related and more likely to correspond to
an admixture event. Figure 6:
Derived origin of admixture for Taiwan populations using TreeMix. In the first stage of this work,
we examined the genetic variability using mitochondrial nucleotide positions
(nps) 8000 to 9000 and nps 10,000 to 11,000 of the coding regions, and the
HVS-I segment of the D-loop region to determine the genetic diversity and
origin of the Saisiyat tribe of Taiwan. In the second stage, we investigated
the general believed that Negrito groups predating the Lower Glacial Maximum
period were associated with the first settlers of Sunderland/Island Southeast
Asia (ISEA) [9]. Although there are some Chinese
accounts of small, dark-skinned people with curly hair in Taiwan, to date,
Taiwan has no archeological human remains supporting this hypothesis.
Interestingly, most Taiwan Mountain tribes have kept folk tales describing past
contacts with Negrito groups, most particularly the Saisiyat tribe of Hsinchu
and Miaoli (Figure 1) who still perform solemn rites to commemorate this past.
On the other hand, the presence of Negrito is still existent in the Philippines
and other parts of peninsular East Asia, but not in Taiwan. If there is any
truth in these TwMtA folk tales, then a small number of Negrito in Taiwan must
have interacted with Neolithic agriculturalist migrants from the Southeast
Asian mainland and most likely shaped their genetic diversity. In this regard,
it is sensible to expect that these early contacts must have left some genetic
traces. Here, our analysis of mtDNA polymorphisms tries to provide new insights
into the history of the Saisiyat tribe [31,32]. Genetic
Structure of Saisiyat To investigate the mtDNA
structure of Saisiyat, we first generated a mismatch distribution (Figure 3). A
bimodal curve was obtained, and the two demographic indexes, the SSD test
(SSD=0.008, P<0.001) and the raggedness (r=0.011, P<0.001) were significantly
different to expectation. These results did not support expansion. Further, the
Bayesian Skyline plot (Figure 4) along with previous results from Ko s research
group did not reveal sufficient structure to visualize a recent population
expansion. These results were in contrast with a highly significant Fu s Fs
test for the Saisiyat tribe [22,28,33]. It possible these results are
revealing a sign that the Saisiyat tribe experienced prehistorically a rapid
population growth from an ancestral population with a small effective
population size. Alternatively, the excess of rare mutations indicated by the
significant negative Fu s Fs test could be the result of recent gene flow
introduced by non-indigenous
groups of Taiwan, such as the Minnan and
Hakka. Finally, The BSP indicated an effective population of approximately 1400
women. This number may actually be inflated since our data set was collected by
LM in 2004, but does not conflict with previously published BPSs, nor with the
actual present-day number of Saisiyat women indicated in the last Census from
the Taiwan Council of Indigenous Peoples [1,2,22]. Distribution The
mtDNA composition of the Saisiyat: Out of 21
different mtDNA haplogroups, 15 haplogroups were characterized as northern
TwMtA haplogroups. Four haplogroups, F2a, E1a1a1, M7b1a2a and Y2 (Figure 4),
constituted 75% of the Saisiyat genome. Consistent with other extant Taiwan
Mountain tribes, the Saisiyat tribe 29% of its mtDNA genome was shared with
Non-Taiwan Aborigines (Supplementary Table S1) and were most likely acquired
from migrants of Mainland East Asia in the last 400 years. Most of the remaining haplogroups
were commonly seen throughout Taiwan Northern tribes. Among them, B4a2a, E2b,
M7b1a2a, R9c1a, and Y2 were also seen in the Philippines. They are thought to
represent a plausible signal for a mid-Holocene out-of-Taiwan expansion or a
signal for a bidirectional migration between Taiwan and the Philippines [18,20,22,24,34-37].
The distribution of haplogroup D6 was intriguing. With a coalescence time of
14,390 years BP (Supplementary Table S1 and Figure S1) haplogroup D6 was seen
at low frequency as D6c in East Asia and Southeast Asia (SEA).
The presence of D6a2 as a single individual in the Pazeh plain tribe, in
Saisiyat (1.1%), in Atayal (4.6%) and in the Mamanwa Negrito group of Mindanao
in the Philippines (3.03%) could represent a genetic indicator of a past
Negrito presence in Taiwan. However, a coalescence age estimate of D6a2 (2,626
years BP, CI 0-5,850) makes this supposition doubtful. It is possible the
sharing of D6a2 between Saisiyat/Atayal and the Mamanwa must be the result of
more recent gene flow. It is conceivable that the
bearers of D6a have experienced a bottleneck between 2,600 and 12,000 years BP,
as suggested by the long stretch of nucleotide variation between D6a and D6a2
in their mtDNA genome (Supplementary Table S1andSupplementary
Figure S1). Lastly, mtDNA
haplogroup Y2 is also shared between the Mamanwa and the Saisiyat and
Atayal tribes. Nonetheless, its distribution in the Philippines (Negritos and
non-Negritos) and its coalescence age estimate (5216 years BP, CI: 529-10046),
characterize Y2 as a signature of the Neolithic expansion of Austronesian agriculturists
in insular East Asia rather than a Negrito signature [14,15,17-21,24,34,38,39]. Principal
Component Analysis To characterize population
structure across Saisiyat, other Taiwan groups, and their relationship with
neighboring populations in MSEA and the Philippines, we performed a DAPC
(Figure 5). The first component captured a clear geographical divide between
Austronesian speaking groups and non-TwA groups including Fujian, Minnan, and
Hakka. Component 2 in Figure 5 disclosed a strong affinity between Northern and
Central TwMtA, and between the Philippines and the Southern TwMtA, suggesting
that all these Austronesian-speaking populations have a common origin. However,
the mtDNA composition of the Saisiyat suggested evidence supporting genetic
affinity between Saisiyat and Negrito and non-Negrito groups of the
Philippines. Using Delfin mtDNA and our Taiwan
data set we constructed a Multidimensional scaling plot to establish this
relationship (Supplementary Figure S3). While the relationship between
populations was the same as in the DAPC plot, the Aeta and Agta Negrito groups
were outbound, most likely because of the conjoint results of drift, the
presence of high frequency haplogroups such as P and M52 in Aeta and Agta, and
long isolation [14]. On the other hand, haplogroup B4b1a2, E1a1a1, Y2, and D6a
in the Mamawan group inferred strong affinity of the Mamanwa Negritos with
other Austronesian groups, suggesting a more recent gene-flow of
Austronesian-speakers in the Mamawa. Maximum
Likelihood Tree from TreeMix TreeMix results inferred mtDNA
gene-flow events (Figure 6) potentially summarizing patterns of population in
the history of Taiwan such as bottlenecks, isolation, consanguinity within
populations, here Saisiyat was characterized as a northern Taiwan tribe.
Further, the gene-flow from Saisiyat (or the Northern TwMtA) to the Central
TwMtA (Bunun, Thao, and Tsou) was previously foreseen in Figure 4 and 5, and is
confirmed in Figure 6 [30]. Moreover, the strong migration event depicted by
TreeMix from the Philippines to Saisiyat indicates genetic interflow. The mtDNA
haplogroups possibly associated with this event, and seen in Saisiyat/Atayal
and the Mamanwa Negrito group of the Philippines, can only be attributed to
subtypes of haplogroups B4b1a2, E1a1a1, Y2, and D6a2. These findings substantiate a
possible past existence of Negritos in Taiwan. They suggest that the Mamanwa
are intermediate between Austronesian and Negritos (Supplementary Figure S3) and possibly experienced several admixture
events in the past. This option is nonetheless not supported by the age
estimate of molecular variation obtained for any of the haplogroups of the same
clade between Mamanwa and Saisiyat/Atayal. For example, D6a2 dates only to 2600
± 1500 yrs BP (Supplementary figure S1) and Y2 dates 3956 ± 2455 yrs BP (Supplementary
figure S2). One possible way to demonstrate a Negrito ancestry in Taiwan
associated to D6 or Y2 would be to find sister branches of these haplogroups in
Taiwan and/or the Philippines that would allow a coalescent node in the
pre-Holocene period. Other
Gene Systems Supporting this last observation,
our previous Y chromosome analysis observed 4 Y-chromosome
single nucleotide polymorphism (Y-SNP)
haplogroups out of 24 unrelated Saisiyat males (Supplementary Table S1). Only
one major haplogroup O1a1* (P203) had a frequency of 87.5%, while
all other haplogroups (O1a2 (M50/110, O3a1* (KL1/122) and O3a2c1a
(M133/M7/M134) were seen only once (4.2%) [21]. When compared to the Y
haplogroup profile of the Philippines, O1a1* (P203) was prevalent in
all Filipino
ethnolinguistisc groups, Negritos and non-Negrito and
its presence in any Negrito groups was regarded as an admixture with the
former. Most interestingly, Negrito groups in the Philippines invariably
possessed, to various levels, haplogroup haplogroups C-RPS4Y/M216, K-M9, and
O3-M122 [15]. These haplogroups have not been seen in Saisiyat, but single
observations of C-RPS4Y/M216 and K-M9 have been seen in the Taiwan plain tribes
and could support a past presence of Negrito in Taiwan [15,21]. Lastly, to our knowledge, no previous
studies associating the Filipino Negrito groups and the HLA gene system have
yet been published; accordingly, no Negrito HLA inferences could be used for
Taiwan. Nonetheless, several HLA*A-B-DRB1 haplotypes were unique to
Saisiyat and 1/3 of Saisiyat haplotypes were shared with Atayal (Supplementary
Table S1). Lastly, the sharing of
haplotype HLA A*34:01-B*56:01 or simply the sole presence
of A*34:01 between Amis, Papua New Guinea highlanders, Maoris of New
Zealand, and Australian Aborigines is intriguing. These findings suggest that
HLA A*34:01 could be a genetic indicator of the pre-dispersal period
of the Negrito throughout ISEA in the late Pleistocene era and should warrant
further HLA analysis of the Philippines Negritos [3,6,40,41]. This investigation has
contributed substantially more insights into the population groups in Taiwan
and the Philippines. Further, while the physical appearance of Negritos has
never been seen in Taiwan, few Taiwan Mountain tribes, such as the Saisiyat and
the Atayal tribes, have conserved folktales inferring prehistoric co-habitation
with them, and to this day, still celebrate this period bi-annually. Among the
few mtDNA haplogroups shared between Taiwan Northern tribes and the Mamanwa
Negritos (B4b1a2, E1a1a1, Y2 and D6a) only D6a may represent a common Negrito
genetic legacy of the Saisiyat and Atayal tribes. This finding must be taken
with caution, as the mid-Neolithic coalescence age estimate of D6a is too
shallow. Further, no support was given from the Y chromosome analysis for
Saisiyat and Atayal. Although the apparent affinity between the Taiwan Northern
tribes and the Mamanwa Negritos of the southern Philippines could be the result
of gene flow brought upon by bidirectional population movements at the time of
the out of Taiwan, the presence of C-RPS4Y/M216 and K-M9 in Taiwan were scarce,
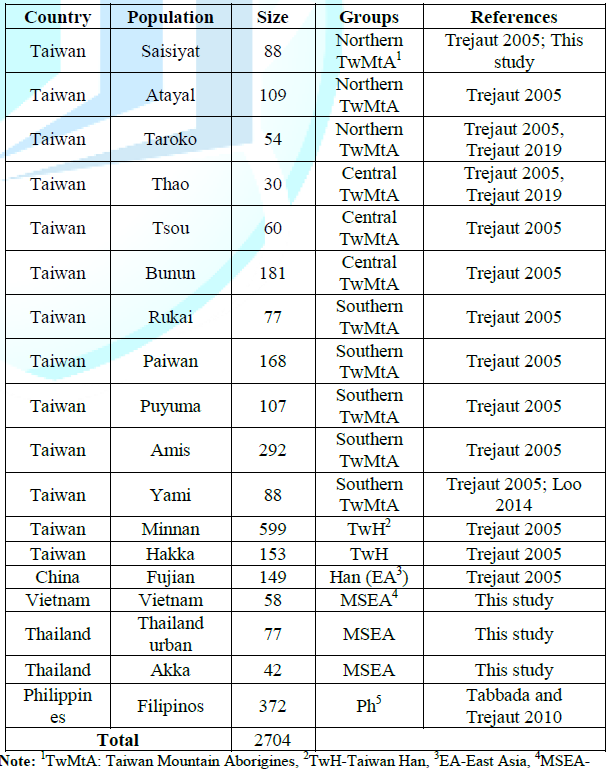
and warrant more extensive studies of the Taiwan gene pool in the future. Samples Whole blood or saliva specimens
were collected from 2704 unrelated individuals (Table 2) comprising Austronesian speaking groups from the
Philippines (n=372), 251 TwMtA consisting of Saisiyat (n=88), Atayal (n=109 )
and Taroko (n=54), 271 central TwMtA consisting of Thao (n=30), Tsou (n=60) and
Bunun (n=181), and 732 southern TwMtA consisting of Rukai (n=77), Paiwan
(n=168), Puyuma (n=107), Amis (n=92) and the Yami islanders (n=88). The
sampling also included 752 Taiwanese of Han descent (TwH) namely Minnan (n=599)
and Hakka (n=153). Samples from neighbor populations included Han individuals
from the east coast of China (Fujian, n=149), groups from Mainland Southeast
Asia (MSEA, n=177), namely Vietnam (n=58), Thailand (n=77) and Akka (n=42), and
finally 372 individuals from the Philippines as described in Tabbada. All samples
above were collected from volunteers with individual written informed consent
during the period of 2001 to 2004 by ML under approval of the Ethics Committee
of the Mackay memorial hospital after giving information regarding the origins
of their parents and grandparents [3,6,20]. Data
Analysis All collected samples in our
dataset were typed for Human
Leukocyte Antigens (HLA-A, -B and -DRB1) and described in the
Anthropology/HLA diversity component of the 13th international histocompatibility
workshop. Specimen typed for mtDNA had haplogroups assigned according to Build
17 of Phylotree. Y haplogroups of the non-recombining part of the Y-chromosome
(NRY) were determined using 81 Y-SNPs, and further analyzed using 17 Y-chromosome
short tandem repeats (Y-STRs): DYS19, DYS385I, DYS385II, DYS389I, DYS389II,
DYSS390, DYS391, DYS392, DYS393, DYS437, DYS438, DYS439, DYS448, DYS456,
DYS458, DYS635, and Y GATA-H4. Additionally, we used corresponding mtDNA data
from the literature including 24 complete mtDNA genome sequences representing
Saisiyat [20-22,24,42,43].The partial mtDNA sequences are available in (Supplementary
Table S2) The whole-mtDNA genome sequencing is available in Supplementary Text
File S1. We deposited 2 new whole- mtDNA sequences in GenBank. Statistical
Analysis In order to test for past
population expansion of Saisiyat, we used two statistical tests Tajima s D and
Fu s Fs [33,44]. The analyses were implemented in Arlequin 3.5.2.2, and
p-values were generated using 1,000 simulations under a model of selective
neutrality [45]. In addition, a mismatch frequency
graph was plotted by using the mtDNA data from Arlequin 3.5.2.2 to determine
whether the population of Saisiyat exhibited evidence of spatial range
expansion or a stationary population history [44]. Demographic variation
through time was obtained from a BSP using Beast with a relaxed molecular clock
and a mutation rate of the mtDNA HVS-I data of 2.2964 x 10-7
mutations per site per year. Adegenet for R was used to perform DAPC with a
number of Principal Components set to 273. The DAPC plot and inertia ellipses
were produced using the poppr module of the R package. A maximum likelihood
tree using mtDNA SNP frequencies was inferred with the TreeMix software.
Admixture and direction of gene flow were inferred using the 20 most
significant events [29,30,46-48]. We are grateful to the people of
Taiwan for donating their blood. The project was conceived and
designed by JAT. JAT and LRC drafted the manuscript equally. LRC performed data
analysis. The laboratory work was performed by ZSC and YHL. All authors have
read and approved the final version of the manuscript. 1.
Council
of Indigenous Peoples, Aboriginal population statistics.
2.
Digital
Museum of Taiwan Indigenous People. 3.
Chu CC, Lin M, Nakajima F, Lee
HL, Chang SL, et al. Diversity of HLA among Taiwan s indigenous tribes and the
Ivatans in the Philippines (2001) Tissue Antigens 58: 9-18. https://doi.org/10.1034/j.1399-0039.2001.580102.x
4.
Chu CC, Trejaut J, Lee H, Chang S
and Lin M. Anthropology/human genetic diversity population reports (2006) 13th
International Histocompatibility Workshop Anthropology/Human Genetic Diversity
Joint Report, USA, pg no. 611-615. 5.
Lin M, Chu C-C, Broadberry R, Yu
L-C, Loo J-H, Trejaut J. Genetic diversity of Taiwan s indigenous peoples:
possible relationship with insular Southeast Asia (2005) In: Sagart, L, Blench
R, Sanchez-Mazas A (Eds) The Peopling of East Asia: Putting Together
Archaeology, Linguistics and Genetics, Routledge Curzon, London and New York,
pg. 230-247. 6.
Lin M, Chu CC,
Chang SL, Lee HL, Loo JH, et al. The origin of Minnan and Hakka, the so-called Taiwanese
, inferred by HLA study (2001) Tissue Antigens 57: 192-199. 7.
Li PJk. A Comparative Vocabulary
of Saisiyat Dialects. Bulletin of the Institute of History and Philology (1978)
Academia Sinica 49: 133-199. 8.
Hu CY. Embodied Memories and
Enacted Ritual Materials-Possessing the Past in Making and Remaking Saisiyat
Identity in Taiwan (2006) United Kingdom. 9.
Bulbeck D. Craniodental
affinities of Southeast Asia s negritos and the concordance with their genetic
affinities (2013) Hum Biol 85: 95-133. https://doi.org/10.3378/027.085.0305
10.
Liu YL. The Study of the Legend
of Pygmy from Taiwanese Indigenous Tribes (2015) China. 11.
Stock JT. The Skeletal Phenotype
of Negritos from the Andaman Islands and Philippines Relative to Global
Variation among Hunter-Gatherers (2013) Human Biolog 85: 67-94. https://doi.org/10.3378/027.085.0304
12.
Migliano AB, Vinicius L and Lahr
MM. Life history trade-offs explain the evolution of human pygmies (2007) Proc
Natl Acad Sci, USA, 104: 20216-20219. https://doi.org/10.1073/pnas.0708024105 13.
Omoto K. The Negritos: genetic
origins and microevolution (1984) Acta Anthropogenet 8: 137-147. 14.
Delfin F, Min-Shan Ko A, Li M,
Gunnarsdottir ED, Tabbada KA, et al. Complete mtDNA genomes of Filipino ethnolinguistic
groups: a melting pot of recent and ancient lineages in the Asia-Pacific region
(2014) Eur J Hum Genet 22: 228-237. https://doi.org/10.1038/ejhg.2013.122
15.
Delfin F, Salvador JM, Calacal GC,
Perdigon HB, Tabbada KA, et al. The Y-chromosome landscape of the Philippines:
extensive heterogeneity and varying genetic affinities of Negrito and
non-Negrito groups (2010) Eur J Hum Genet 19: 224-230. https://doi.org/10.1038/ejhg.2010.162
16.
Migliano AB, Romero IG, Metspalu
M, Leavesley M, Pagani L, et al. Evolution of the pygmy phenotype: evidence of
positive selection from genome-wide scans in African, Asian, and Melanesian
pygmies (2013) Hum Biol 85: 251-284. https://doi.org/10.3378/027.085.0313
17.
Karafet TM, Osipova LP, Gubina
MA, Posukh OL, Zegura SL, et al. High levels of Y-chromosome differentiation
among native Siberian populations and the genetic signature of a boreal
hunter-gatherer way of life (2002) Hum Biol 74: 761-789. https://doi.org/10.1353/hub.2003.0006
18.
Soares P, Trejaut JA, Loo JH,
Hill C, Mormina M, et al. Climate change and postglacial human dispersals in
southeast Asia (2008) Mol Biol Evol 25: 1209-1218. https://doi.org/10.1093/molbev/msn068
19.
Soares PA, Trejaut JA, Rito T,
Cavadas B, Hill C, et al. Resolving the ancestry of Austronesian-speaking
populations (2016) Hum Genet 135: 309-326. https://doi.org/10.1007/s00439-015-1620-z
20.
Tabbada KA, Trejaut J, Loo JH,
Chen YM, Lin M, et al. Philippine mitochondrial DNA diversity: a populated
viaduct between Taiwan and Indonesia? (2010) Mol Biol Evol 27: 21-31. https://doi.org/10.1093/molbev/msp215
21.
Trejaut JA, Poloni ES, Yen JC,
Lai YH, Loo JH, et al. Taiwan Y-chromosomal DNA variation and its relationship
with Island Southeast Asia (2014) BMC Genet 15: 77. https://doi.org/10.1186/1471-2156-15-77
22.
Ko AM, Chen CY, Fu Q, Delfin F,
Li M, et al. Early Austronesians: into and out of Taiwan (2014) Am J Hum Genet
94: 426-436. https://doi.org/10.1016/j.ajhg.2014.02.003
23.
Tajima A, Sun CS, Pan IH, Ishida
T, Saitou N, et al. Mitochondrial DNA polymorphisms in nine aboriginal groups
of Taiwan: implications for the population history of aboriginal Taiwanese
(2003) Hum Genet 113: 24-33. https://doi.org/10.1007/s00439-003-0945-1
24.
Trejaut JA, Kivisild T, Loo JH,
Lee CL, He CL, et al. Traces of archaic mitochondrial lineages persist in
Austronesian-speaking Formosan populations (2005) PLoS Biol 3: e376. https://doi.org/10.1371/journal.pbio.0030376
25.
Soares P, Ermini L, Thomson N,
Mormina M, Rito T, et al. Correcting for purifying selection: an improved human
mitochondrial molecular clock (2009) Am J Hum Genet 84: 740-759. https://doi.org/10.1016/j.ajhg.2009.05.001
26.
Anderson SAT, Bankier BG, Barrell
BG, de Bruijn MH, Coulson AR, et al. Sequence and Organization of the Human
Mitochondrial Genome (1981) Nature 290: 457-465. https://doi.org/10.1038/290457a0
27.
Nei M. Molecular evolutionary
genetics (1987) Columbia University Press, United States. 28.
Harpending HC. Signature of
ancient population growth in a low-resolution mitochondrial DNA mismatch
distribution (1994) Hum Biol 66: 591-600. 29.
Drummond AJ and Rambaut A. BEAST:
Bayesian evolutionary analysis by sampling trees (2007) BMC Evol Biol 7: 214. https://doi.org/10.1186/1471-2148-7-214
30.
Pickrell JK and Pritchard JK.
Inference of population splits and mixtures from genome-wide allele frequency
data (2012) PLoS Genet 8: e1002967. https://doi.org/10.1371/journal.pgen.1002967
31.
Quartly J. In honor of the little
black people (2004) Taipei Times. 32.
Endicott P. Revisiting the Negrito
Hypothesis: A Transdisciplinary Approach to Human Prehistory in Southeast Asia
(2013) Hum Biol 85: 7-20. https://doi.org/10.3378/027.085.0301
33.
Fu YX. Statistical tests of
neutrality of mutations against population growth, hitchhiking and background
selection (1997) Genetics 147: 915-925. 34.
Brandao A, Eng KK, Rito T,
Cavadas B, Bulbeck D, et al. Quantifying the legacy of the Chinese Neolithic on
the maternal genetic heritage of Taiwan and Island Southeast Asia (2016) Hum
Genet 135: 363-376. https://doi.org/10.1007/s00439-016-1653-y
35.
Bellwood P. In the origin and
dispersals and dispersal of agricultural communities in Southest Asia, Glover I
and Bellwood P (Ed) (2004) Taylor and Francis group, United Kingdom, pg
no.21-40. 36.
Diamond J and Bellwood P. Farmers
and their Languages: the first expansions (2003) Science 300: 597-603. https://doi.org/10.1126/science.1078208 37.
Loo J, Trejaut J, Yen J, Chen Z,
Lee C, et al. Genetic affinities between the Yami tribe people of Orchid Island
and the Philippine Islanders of the Batanes archipelago (2011) BMC Genet 12: 21.
https://doi.org/10.1186/1471-2156-12-21
38.
Behar DM, van Oven M, Rosset S,
Metspalu M, Loogväli EL, et al. A Copernican reassessment of the human
mitochondrial DNA tree from its root (2012) Am J Hum Genet 90: 675-684. https://doi.org/10.1016/j.ajhg.2012.03.002
39.
Hill C, Soares P, Mormina M,
Macaulay V, Clarke D, et al. A mitochondrial stratigraphy for island southeast
Asia (2007) Am J Hum Genet 80: 29-43. https://doi.org/10.1086/510412
40.
Chu CC, Lee HL, Trejaut J, Chang
HL and Lin M. HLA-A, -B, -Cw and -DRB1 allele frequencies in a Pazeh population
from Taiwan Fernandes-Vina (2004) Human Immunol 65: 1102-1181. https://doi.org/10.1016/j.humimm.2004.08.140
41.
Single RS, Meyer D, Mack SJ,
lancaster A, Nelson MP, et al. Immunobiology of the Human MHC (2002) 13th
International Histocompatibility Workshop Anthropology/Human Genetic Diversity
Joint Report, United States, pg no.705-746. 42.
Middleton D, Menchaca L, Rood H
and Komerofsky R. New allele frequency database (2003) Tissue Antigens 61:
403-407. https://doi.org/10.1034/j.1399-0039.2003.00062.x
43.
Van Oven M and Kayser M. Updated
comprehensive phylogenetic tree of global human mitochondrial DNA variation
(2009) Hum Mutat 30: E386-394. https://doi.org/10.1002/humu.20921
44.
Tajima F. Statistical method for
testing the neutral mutation hypothesis by DNA polymorphism (1989) Genetics
123: 585-595. 45.
Excoffier L, Laval G and
Schneider S. Arlequin (version 3.0): an integrated software package for
population genetics data analysis (2007) Evol Bioinform Online 1: 47-50. https://doi.org/10.1177/117693430500100003
46.
Jombart T. Adegenet: A R Package
for the Multivariate Analysis of Genetic Markers (2008) Bioinformatics 24:
1403-1405. https://doi.org/10.1093/bioinformatics/btn129
47.
R Development core team. A
language and environment for statistical computing. R foundation for
statistical computing (2013) Vienna, Austria. Kamvar ZN, Tabima JF and Grünwald NJ. Poppr: an
R package for genetic analysis of populations with clonal, partially clonal,
and/or sexual reproduction (2014) Peer J 2: e281. https://doi.org/10.7717/peerj.281 Jean
Alain Trejaut, Molecular Anthropology and Transfusion Medicine Research
Laboratory, Mackay Memorial Hospital, #45, Min-Sheng Road, Tamsui, New Taipei
City, 25115, Taiwan, Tel: +886-2-2809-4661, Fax:
+886-2-2809-8746, E-mail: jtrejaut@gmail.com Chen
LR, Trejaut JA, Lai YH, Chen ZS, Huang JY, et al. Mitochondrial DNA polymorphisms
of the saisiyat Indigenious group of Taiwan, search for a negrito signature (2019)
Edel
J Biomed Res Rev 1: 12-18. Molecular Anthropology, Negrito, Austronesian,
Population genetics, Saisiyat, Taiwan aborigines, Mitochondrial DNA. Mitochondrial DNA Polymorphisms of the Saisiyat Indigenous Group of Taiwan, Search for a Negrito Signature
Abstract
Full-Text
Introduction
Results
Discussion
Summary
Material
and Method
Acknowledgements
Authors contributions
References
*Corresponding authorCitation
Keywords