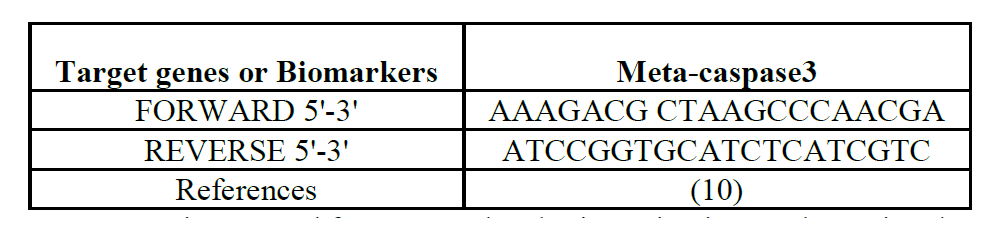
Research Article :
Spondias mombin is a
plant that has been traditionally noted for its medicinal with a preliminary
results report a wide range of antibacterial and antifungal properties.
Meta-caspases and Caspases are essential in cells for programmed cell death, in
development and most other stages of adult life, and have been termed
"executioner" proteins for their roles in the cell. A 12 hours old
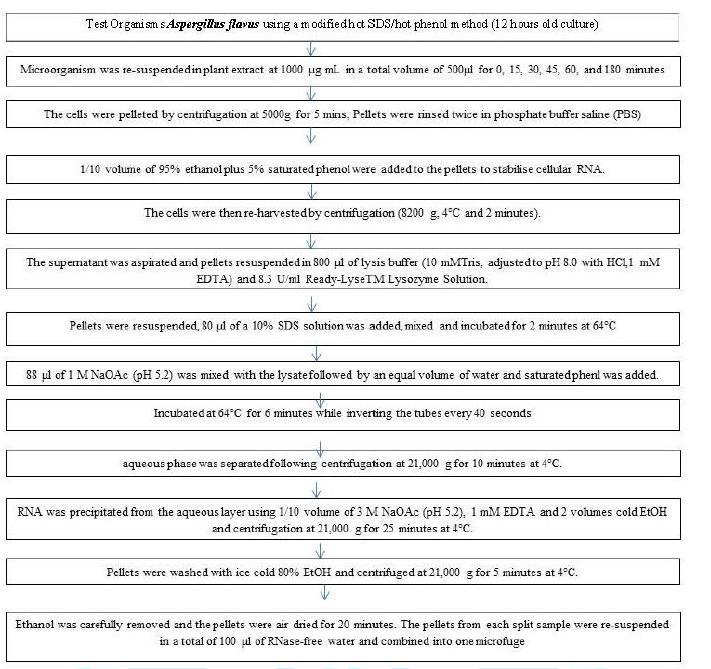
culture of each microorganism was re-suspended in plant extract at 1000 µg mL
in a total volume of 500 µl for 0, 15, 30, 45, 60, and 180 minutes. The cells
were pelleted by centrifugation at 5000 g for 5 minutes. The pellets were
rinsed twice in phosphate buffer saline (PBS). Then 1/10 volume of 95% ethanol
plus 5% saturated phenol were added to the pellets to stabilize cellular RNA.
The cells were then re-harvested by centrifugation (8200 g, 4°C and 2 minutes).
The supernatant was aspirated and pellets re-suspended in 800 μl of lysis
buffer (10 mMTris, adjusted to pH 8.0 with HCl, 1 mM EDTA) and 8.3 U/ml
Ready-LyseTM Lysozyme Solution. After the pellets were re-suspended, 80 μl of a
10% SDS solution was added, mixed and incubated for 2 minutes at 64 °C. Then 88
μl of 1 M NaOAc (pH 5.2) was mixed with the lysate followed by an equal volume
of water and saturated phenol was added. Total RNA was quantified using Spectrophotometric
absorbance at 260 nm DNA was removed with Turbo DNA-free (Ambion, Inc.).
Reverse Transcription-PCR reaction was performed in a 15.0 µl final volume (kit
number-DNA-PCR739288). Assessment of Polymerase Chain Reaction products
(amplicons) were electrophoreses in 0.5% of agarose gel using 0.5 × TBE buffer
( 2.6 g of Tris base, 5 g of Tris boric acid and 2 ml of 0.5M EDTA and adjusted
to pH 8.3 with the sodium hydroxide pellet) with 0.5 μl ethidum bromide. The
mechanism of action of isolated novel compounds using Metacaspase3 to programme
the death of test organism (Aspergillus flavus) between 0 and 180 minutes
interval. It was observed that cell (via DNA) were completely destroyed at 180
minutes with all the isolated compounds. The purpose of this research work is
to evaluate the programmed cell death (PCD) of Aspergillus flavus by
triggered Cysteine-dependent Aspartate-directed proteases (meta-caspase3)
lethality mechanism of novel compound isolated from ethyl acetate extract
ofSpondias mombin. Spondias mombin is a plant that has been traditionally noted
for its medicinal and food values. Preliminary results report a wide range of
antibacterial and antifungal properties [1, 2]. Scientific investigations have
shown that it has anthelmintic, antioxidant, antimicrobial and anti-
inflammatory actions [3, 4]. Meta-caspases can be defined as cysteine-dependent
aspartate-directed proteases are a family of cysteine proteases that play
essential roles in apoptosis (programmed cell death), necrosis, and inflammation.
Caspases are essential in cells for apoptosis, or programmed cell death, in
development and most other stages of adult life, and have been termed
"executioner" proteins for their roles in the cell. Some caspases are
also required in the immune system for the maturation of lymphocytes. Failure
of apoptosis is one of the main contributions to tumor development and
autoimmune diseases this, coupled with
the unwanted apoptosis that occurs with ischemia or Alzheimers disease, has
stimulated interest in caspases as potential
therapeutic targets. Inactive protease of caspase family is in state of
pro-enzyme which amino acid end has a sequence called “pro-domain”. When
zymogen is being activated, the pro-domain is cleaved and the rest part is cut
into two subunits called P20 and P10. Active zymogens consist of these two
subunits in forms of (P20/P10)2. This activation reaction is also Asp-specific
for reason that the cleavage occurs between Asp of conserved sequences in
pro-enzyme and the amino acids sequence after Asp. In the cleavage the small
subunit at the carboxyl end is cleaved first and pro-domain is then cut off the
amino side of the big subunit. The cleavage can be self-catalyzing of
pro-enzyme and mediated enzyme or functioned by other proteases of ICE family. Pro-caspase-3 has 277 amino acids, molecular weight of 32kD,
30% homology with ICE and 35% homology with CED-3. In caspase family
pro-caspase-3 is the most homologous to CED-3 both in structure and substrate
specificity. The pro-domain of caspase-3 is shorter than that of ICE which has
28 amino acids, but its activity center and conserved amino acids that are
related with substrate binding are the same with ICE. In activation,
pro-caspase-3 is cleaved at two sites: Asp28~Ser29 and Asp175~Ser176, giving
rise to two fragments: P17 (29~175) and P10 (182~277) which are close to P20
and P10 of ICE. The two subunits combine and form active caspase-3. When being
activated, pro-caspase-3 is not active of catalyzing before being cleaved by
granzyme B (GrzB) or caspase-10 at D175. Other caspases e.g. ICE might
participate in cleaving pro-domain of caspase-32.0 [4]. Caspase-3 is indispensible in apoptosis. It triggers
apoptosis when being transfected into insect Sf9 cells. This process can be
blocked by BCL-2. Exclusion of caspase-3 in extractions of apoptotic cells
leads to loss of capability of inducing apoptosis. Adding of caspase-3 let it
regain the capability of inducing apoptosis. Caspase-3 can be activated by
various factors. In CTL-mediated killing, caspase-3 can be activated both by
Fas/FasL pathway and by granzyme B pathway. Granzyme B is a kind of serine
esterase in cells, and is the only protease that cleaves after Asp except
caspases in mammals. Granzyme B can specifically cleave IxxD sequence at the C
terminal of catalyzing subunit of ICE family and activate caspase-2, 3, 6, 7,
8, 9, 10. ICE can be cleaved by granzyme B, too, but it cant be activated after
cleavage. The purpose of this research work is to evaluate the Programmed
Cell Death (PCD) of Aspergillus flavus by triggered Cysteine-dependent
Aspartate-directed proteases (meta-caspase3) lethality mechanism of novel
compound isolated from ethyl acetate extract of Spondias mombin. Microorganism for the research work The strains used for this research work were fungi collected
from Central Medical Laboratory (CML), Obafemi Awolowo University Teaching,
Hospital (OAUTH), Ile Ife, Osun State, and the Institute of Advance Medical
Research and Training (IMRAT), University College Hospital, Ibadan, Oyo State
Nigeria. Sources of microorganisms The strains used for this research work were fungi collected
from Central Medical Laboratory (CML), Obafemi Awolowo University Teaching,
Hospital (OAUTH), Ile Ife, Osun State, and the Institute of Advance Medical
Research and Training (IMRAT), University College Hospital, Ibadan, Oyo State
Nigeria. The identity of the test organisms was confirmed ID 32 C
system (Biomerieux, France) following the manufacturers instructions. The yeast
isolates were identified by the ID 32 C Analytical Profile Index [6]. A 12 hours old culture of each microorganism was
re-suspended in plant extract at 1000 µg mL in a total volume of 500 µl for 0,
15, 30, 45, 60, and 180 minutes. The cells were pelleted by centrifugation at
5000 g for 5 minutes. The pellets were rinsed twice in phosphate buffer saline
(PBS). Then 1/10 volume of 95% ethanol plus 5% saturated phenol were added to
the pellets to stabilise cellular RNA. The cells were then re-harvested by
centrifugation (8200 g, 4 °C and 2 minutes). The supernatant was aspirated and
pellets re-suspended in 800 μl of lysis buffer (10 mMTris, adjusted to pH 8.0
with HCl, 1 mM EDTA) and 8.3 U/ml Ready-LyseTM Lysozyme Solution. After the
pellets were re-suspended, 80 μl of a 10% SDS solution was added, mixed and
incubated for 2 minutes at 64 °C. Then 88 μl of 1 M NaOAc (pH 5.2) was mixed
with the lysate followed by an equal volume of water and saturated phenol was
added. This was incubated at 64°C for 6 minutes while inverting the tubes every
40 seconds. The aqueous
phase was separated following centrifugation at 21, 000 g for 10 minutes at
4°C. The RNA was precipitated from the aqueous layer using 1/10
volume of 3 M NaOAc (pH 5.2), 1 mM EDTA and 2 volumes cold EtOH and
centrifugation at 21, 000 g for 25 minutes at 4°C. Pellets were washed with ice
cold 80% EtOH and centrifuged at 21, 000 g for 5 minutes at 4°C. The ethanol
was carefully removed and the pellets were air dried for 20 minutes. The
pellets from each split sample were re-suspended in a total of 100 μl of
RNase-free water and combined into one microfuge [7] (kit
number-DNA-PCR739288). Total RNA was quantified using spectrophotometric absorbance
at 260 nm DNA was removed with Turbo DNA-free (Ambion, Inc.). Removal of DNA
from the RNA samples was performed using DNA-free™ DNA Removal Kit
(ThermoFisher) following manufacturers protocol. Purified DNA-free RNA was
converted to cDNA immediately using ProtoScript® First Strand cDNA Synthesis
Kit (NEB). The cDNA was diluted to a final volume of 286 μl and stored at 4°C
(8). PCR protocol Reverse Transcription-PCR reaction was performed in a 15.0
µl final volume. Briefly, 1 µl template cDNA (40 ng) was combined with 1.0 µl
of forward primer (5 nM), 1.0 µl of reverse primer (5 nM), 4.5 ml nuclease-free
water and 7.5 µl of Taq 2X Master Mix. Thermo cycling was performed by 40
cycles at 95 °C for 15 seconds, 60 °C for 15 seconds and 72 °C for 15 seconds.
Analysis of the PCR products was performed using 1.5% agarose gel solution in
TBE buffer and visualisation was enabled by soaking gel in ethidium bromide
solution for 10 minutes and UV-transilluminator. The data obtained were analyed
using Graph pad prism version 6.01 description and frequency. statistic was
generated to describe the diameter of inhibition. quantitative phytochemical
constituent and toxicological prameter to test for the level of significance
[8]. Gel electrophoresis Assessment of Polymerase Chain Reaction
products (amplicons) were electrophoresed in 0.5% of agarose gel using 0.5
× TBE buffer (2.6 g of Tris base, 5 g of Tris boric acid and 2 ml of 0.5M EDTA
and adjusted to pH 8.3 with the sodium hydroxide pellet) with 0.5 μlethidum
bromide. The expression product was visualized as bands by UV-transilluminator
[8-10] (Table 1 & Chart 1) Chart 1: Isolation of RNA from bacterial cell. Programmed Cell Death of Aspergillus flavus Trigger
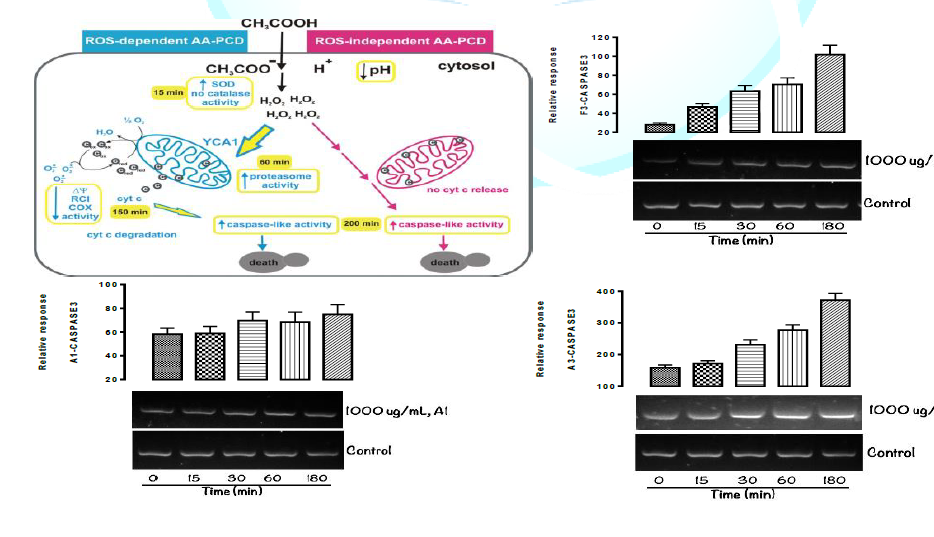
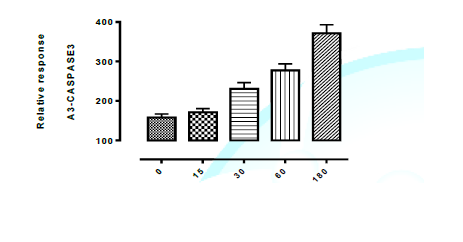
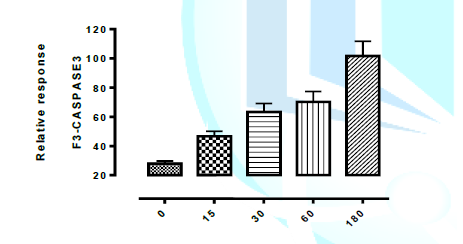
Meta-caspase3 Lethality mechanism. Figure 1 shows the mechanism of action of
isolated novel compounds using Metacaspase3 to programme the death of test
organism (Aspergillus flavus) between 0 and 180 minutes interval. It was
observed that cell (via DNA) were completely destroyed at 180 minutes with all
the isolated compounds from Spondias mombin. The graphically represented in
Figures 1, 1a, 1b, and 1c of isolated novel compounds and Programmed Cell Death
in Aspergillus flavus trigger meta-caspase3 Lethality by Aspergillus flavus
were demonstrated below. It can be deduced that the isolated novel compounds
has fungicidal mechanism of action using triggered Meta-caspase3 lethality to
program the death of the test fungus (Aspergillus flavus). The purpose of this research work is to evaluate the
Programmed Cell Death (PCD) of Aspergillus flavus by triggered
Cysteine-dependent Aspartate-directed proteases (meta-caspase3) lethality
mechanism of novel compound isolated from ethyl acetate extract of Spondias
mombin. The isolated compound are A1- Epigallocatechin, Epicatechin and
Stigmasterol phytosterol (Synergy), A3-Aspidofractinine-3-methanol and
F3-Terephthalic dodecyl 2-ethylhexyl ester. Apoptosis or Programmed Cell Death
(PCD) is a prominent feature of a developing cell signalling. Metacaspase3 is a
caspase protein that interacts with caspase-8 and caspase-9. It is a protein
and member of cysteine-aspartic acid protease. Sequential activation of caspase
plays a central role in the execution phase of cell apoptosis [11]. In this
present study, the mechanism of Caspase3 was discussed as mention by previous
research [12]. It was reported that Caspase involved the catalytic site and
sulfohydyl group of cys-285 and the imidazole ring of His 237-His 237
stabilizes the carbonyl group of the key aspartate residue while cys 285
attacks to ultimately cleaves the peptide bond. Cys 285 and Gily 238 also
function to stabilize the tetrahedral transition state of the substrate enzyme
complex through hydrogen bonding [13]. In figure 1, 1a, 1b and 1c the breakdown of the pathway
mechanism are as follow. Amino Acid (AA) enters Aspergillus flavus cells by
diffusion through the plasma membrane. In the cytosol, AA dissociates into
Acetate and protons causing intracellular acidification. Alternative PCD
pathways are induced by Amino acid (AA): a ROS-dependent (blue lines) and a
ROS-independent (pink lines) pathway. H2O2 accumulates early in both the
pathways. In the ROS-dependent
pathway SOD activity increases at 15 min. YCA1 acts upstream of cyt c (c)
release from mitochondria to the cytosol released cyt c acts as an electron donor
(cred) to mitochondrial respiratory chain and as superoxide anion (O2•−)
scavenger (cox) and is degraded by unidentified proteases in a late phase mitochondrial functions progressively decline
as judged by decrease in mitochondrial membrane potential, Respiratory Control
Index (RCI) and COX activity caspase-like activity increases in a late
phase with a complete loss of cell viability at 200 min. In the ROS-independent
AA-PCD pathway, cyt c is not released into the cytosol but the caspase-like
activity increases in a late phase (14). In figure 1, Metacaspase3 is activated in the apoptotic cell
both by extrinsic (Death ligand) and intrinsic (mitochondrial pathway) [15].
The mitochondrial pathway was demonstrated in the fig 1. In mitochondrial
pathway, Zymogen features of Caspase3 is necessary, if unregulated Caspase
actively would kill cell indiscriminately. As an executioner Caspase, the
Caspase-3 zymogen has virtually no activity until it is cleared by an initiator
Caspase after apoptotic signalling events like the inclusion of isolated
compound A1, A3 and F3. On inclusion, it can activate initiator Caspases into
cells targeted for apoptosis by killer T cells [14, 15] In Metacaspase3, A1, A2
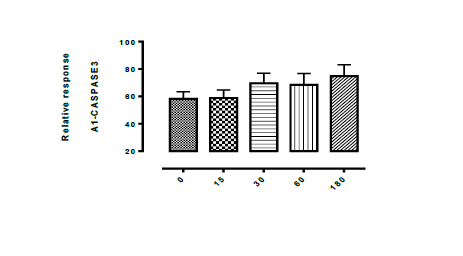
and F3, it was observed that there is a complete inhibition of Aspergillus
flavus (fungi) and their death phase were adequately measured between 0 and 180
minutes interval. The Aspergillus flavus relative response was demonstrated by
the graph in the figure 1, 1a, 1b and 1c, at 0-180 mins interval.it was
observed that major substrate of metacaspase-3 is Poly ADP-ribose polymerase
(PARP) which was found in Aspergillus flavus. It correlates with DNA repairmen
and monitoring of gene integration. It was reported by Boutright and Salvesen
(2003) that metacaspase 3 exist as inactive pro-enzymes that undergo
proteolytic processing at conserved aspartic residues to produce two subunits
large and small that dimerize to form the active enzyme, this protein cleaves
and activates caspase 6 and 7 and two protein itself is processed and activated
by metacaspases 8, 9 and 10. It is the predominant metacaspase involved in the
cleavage of amyloid beta for a precursor protein which is associated with
neuronal death. Alternative splicing of this gene results in the two transcript
variants that encode the same protein [16]. The mechanism of apoptosis is highly complex and involves
energy dependent cascade of molecular events. It is mediated mainly through
three pathways: extrinsic, intrinsic and perforin pathway. The apoptotic mode
of cell death is an active and defined process which plays an important role in
the development of multicellular organisms and in the regulation and
maintenance of the cell populations in tissues upon physiological and
pathological conditions. During the initiation of apoptosis in this study, PARP
116kD is cleaved by metacaspase3 into two fragments, 31kD and 85kD at
Asp216-Gly217, separating its two zinc finger domain that binds with DNA from
its catalyzing domain of carboxyl end, and loses its normal function. Then the
activity of endonuclease which is down regulated by PARP and dependent on Ca2+/Mg2
increases, and DNA in nucleosomes is lysed which triggers apoptosis which can
be observed during the course of the research. This lysis process can be
inhibited by Ac-DEVD-CHO, a specific inhibitor of metacaspase-3, but can’t be
inhibited by CrmA. Metacaspase-3 can also cleave U1-70K、DNA-PK、PKCd
and PKCq. Both PKCd and PKCq belong to novel PKC (nPKC). After being cleaved by
metacaspase-3 which cuts off the regulation domain, they become active PKC.
Moreover, over expression of PKCd and PKCq can trigger apoptosis, which
illustrates they participate in inducing of apoptosis [17]. Sokolov et al reported in vitro metacaspase 3 was found to
prefer the peptide sequence DEXDG (Asp-Glu-Val-Asp-Gly) with cleavage occurring
on the carboxy side of the second aspartic acid residue (between D and G)
metacaspase 3 is active over a broad pH range that is slightly higher (more
basic) than many of the other executioner metacaspases, this broad range
indicates that mtacaspase3 will be fully active under normal and apoptotic cell
condition .It is important to discuss the activation of this programmed cell
death, this has helped to measure the inhibitor factors of isolated compound
A1, A3 and F3 on selected microbe during this research work [18,19]. In Extrintics activation, it will trigger the hallmark
Caspase-cascade characteristic of the apoptotic pathway, in which metacaspase 3
plays a dominant role. It should be noted in the scope of this research work
and reported by previous authors that mitochondria works in combination with
caspase 9, apoptosis-activating factors 1 (Apat 1) and ATP to process
procaspase 3, these molecules are sufficient to activate caspase 3 invitro but
other regulator proteins are necessary in vivo [20]. The major substrate of
metacaspase 3 is poly ADP ribose polymerase (PARP), it correlates with DNA
repairmen, damages and monitoring of gene integration to initiate, all this
were clearly stated in the Figures 1, 1a, 1b and 1c. In conclusion, Cysteine-dependent Aspartate-directed
proteases (meta-caspase3) lethality is the best method to measure the mechanism
of action of lethal compound from spondias mombin isolated compound on microbe
cells and to demonstrate lethality and mechanism of the isolated compound on
the cell of Aspergillus flavus. This is an Alternative to after the early burst
of intracellular H2O2 accumulation, AA-PCD can proceed via a ROS- and
YCA1-independent pathway, in which the death rate is faster than that of the
ROS-dependent pathway, cytc is not released, but still a late caspase-like
activity increase is observed which is not affected by H2O2 scavengers, such as
N-acetyl-L-cysteine, therefore Aspergillus flavus can used to demonstrate
effects of Cysteine-dependent Aspartate-directed proteases (meta-caspase3)
lethality. 1. Okwu DE.
Evalution of chemical composition of indigenous spices and flavouring agents
(2001) Global J Appl Sci 7: 455-459. http://dx.doi.org/10.4314/gjpas.v7i3.16293
2. Abdel-Kader,
M, Hoch J, Berger JM, Evans R, Miller JS, et al. Two bioactive saponins from
Albizia subdimi diata from the Suriname rainforest (2001) J Natural Products
64: 536-539. https://pubs.acs.org/doi/10.1021/np000295u 3. Calderon
AI, Angerhofer CK, Pezzuto JM, Farnsworth NR, Foster R, et al. Forest plots as
a tool to demonstrate the pharmaceutical potential of plants in a tropical
forest of Panama (2010) Econ Bot 53: 278-294.
https://doi.org/10.1007/BF02864782 4. Abulreesh
HH. Multidrug-Resistant Staphylococci in the Environment (2011) International
Conference on Biotechnology and Environment Management IPCBEE 18, IAC SIT
Press, Singapore. https: //scialert.net/abstract/?doi=jm.2011.510.523 5. Oludare
temitope Osuntokun, AO, Oluduro, TO Idowu & AO Omotuyi (2017) Assessment of
Nephrotoxicity, Anti-inflammatory and Antioxidant properties of
Epigallocatechin, Epicatechin and Stigmasterol phytosterol (synergy) Derived
from ethyl acetate stem bark extract of Spondias mombin on Wister Rats Using
Molecular method of analysis. J Mol Microbiol 1: 1-11. 6. Pereira
C, Chaves S, Alves S, Salin B, Camougrand N, et al. (2010) Mitochondrial
degradation in acetic acid-induced yeast apoptosis: the role of Pep4 and the
ADP/ATP carrier. Mol. Microbiol 76: 1398-1410.
https://doi.org/10.1111/j.1365-2958.2010.07122.x 7. Guaragnella
N, Passarella S, Marra E and Giannattasio S. Knock-out of metacaspase and/or
cytochrome c results in the activation of a ROS-independent acetic acid-induced
programmed cell death pathway in yeast (2010) FEBS Lett 584: 3655-3660. https://doi.org/10.1016/j.febslet.2010.07.044
8. Huttemann
M, Pecina P, Rainbolt M, Sanderson TH, Kagan VE, et al. The multiple functions
of cytochrome c and their regulation in life and death decisions of the
mammalian cell: from respiration to apoptosis (2011) Mitochondrion 11: 369-381.
https: //doi.org/10.1016/j.mito.2011.01.010 9. Carmona-Gutierrez
D, Eisenberg T, Buttner S, Meisinger C, Kroemer G, et al. Apoptosis in yeast:
triggers, pathways, subroutines. Cell Death Differ (2010) 17: 763-773.
https://doi.org/10.1038/cdd.2009.219 10. Sokolov S,
Knorre D, Smirnova E, Markova O, Pozniakovsky A, et al. Ysp2 mediates death of
yeast induced by amiodarone or intracellular acidification (2006) Biochim
Biophys Acta 1757: 1366-1370. https://doi.org/10.1016/j.bbabio.2006.07.005 11. Guaragnella
N, Bobba A, Passarella S, Marra E and Giannattasio S. Yeast acetic acid-induced
programmed cell death can occur without cytochrome c release which requires metacaspase
YCA1. (2010) FEBS Lett 584: 224-228. https://doi.org/10.1016/j.febslet.2009.11.072
12. Alberts M
(2008) Molecular Biology of the Cell 5th
Edition, Garland, pp: 34-582. 13. Khurshidi
A, Mahboob H and Nishawar J. Cold resistance in Plants (2008) Department of
Biotechnology, the University of Kashmir, India 342-351. 14. Yobsan
Tamirul, Nigus Abebe and Abriham Kebede. Review on mechanisms of regulating
apoptosis in animal cells, World (2017) J Biomed Pharma Sci 2: 33-38.
https://doi.org/10.5281/zenodo.837832 15. Mohamed A.
Cell Engineering. Zoology (2010) Birmingham University, UK. 16. Osuntokun
OT, Omotuyi OI. Bacteriostatic and Bactericidal Mechanism of Novel Compound
Isolated from Ethyl Acetate Stem Bark Extract of Spondias mombin Using Biomarker
Repressor LexA gene on Escherichia coli and Bacillus subtilis (2018) J Mol
Biomark Diagn 9: 405. https://doi.org/10.4172/2155-9929.1000405 17. Koltover
VK. Antioxidant biomedicine: From free radical chemistry to systems biology
mechanisms (2010) Russi Chem Bull 59: 37-42. http://dx.doi.org/10.31031/RMES.2018.03.000565
18. Babu PV,
Liu D, Gilbert ER (2013) Recent advances in understanding the anti-diabetic
actions of dietary flavonoids. J Nutr Biochem 24: 1777-1789.
https://doi.org/10.1016/j.jnutbio.2013.06.003 19. Kim HS,
Quon MJ, Kim JA (2014) New insights into the mechanisms of polyphenols beyond
antioxidant properties: Lessons from the green tea polyphenol,
Epigallocatechin3-gallate. Redox Biol 2: 187-195.
https://doi.org/10.1016/j.redox.2013.12.022 20. PT Illing,
JP Vivian, Dudek L, Kostenko Z, Chen MB, et al. (2012) Immune self-reactivity
triggered by drug-modified HLA-peptide repertoire. Nature 486: 554-558.
https://doi.org/10.1038/nature11147 Oludare Temitope Osuntokun, Department of
Microbiology, Faculty of Science, Adekunle Ajasin University, Akungba Akoko,
P.M.B 001, Ondo State, Nigeria, E-mail: osuntokun4m@yahoo.com Citation Osuntokun OT, Omotuyi OI, Oluduro AO and Idowu
TO. Assessment of programmed cell death of aspergillus flavus by
triggered cysteine-dependent aspartate-directed proteases (meta-caspase3)
lethality mechanism of novel compounds isolated from ethyl acetate extract
of spondias mombin (2019) Biochem and Modern Appli 2: 30-34 Programmed Cell Death, Aspergillus flavus, Cysteine-dependent Aspartate-directed proteases (Meta-caspase3), Spondias mombin, Novel compound (A1- Epigallocatechin, Epicatechin and Stigmasterol phytosterol (Synergy),A3-Aspidofractinine-3-methanol and F3-Terephthalic dodecyl 2-ethylhexyl ester).
Assessment of Programmed Cell Death of Aspergillus flavus by Triggered Cysteine-dependent Aspartate-directed Proteases (Meta-caspase3) Lethality Mechanism of Novel Compounds Isolated from Ethyl Acetate Extract of Spondias mombin
Abstract
Full-Text
Introduction
Materials and Method
Isolation of RNA
Synthesis of convertible (cDNA)
Results
Discussion
References
Keywords